- 1Department of Botany, Government Degree College, Zhob, Balochistan, Pakistan
- 2Botanical Garden of Osnabrück University, Osnabrück, Germany
- 3National Herbarium, National Agricultural Research Centre, Islamabad, Pakistan
- 4Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany
- 5Department of Botani, Goverment Girls Degree College, Zhob, Balochistan, Pakistan
- 6Department of Agricultural Extension, Zhob, Balochistan, Pakistan
A new species, Allium sulaimanicum, is described from northern Balochistan and southern Khyber Pakhtunkhwa in Pakistan based on morphological, molecular, and cytological studies. The new species is characterised by long runner-like cylindrical rhizomes of adult plants, cylindrical bulbs, linear leaves with minute soft hairs along veins, campanulate perigonium, and white to creamy white, ovate to elliptical, 4.5–5-mm-long acute tepals, with brownish to purplish nerves, stamens as long as to slightly longer than tepals, yellow to brick red anthers, hexagonal ovary, and white and papillate/warty along angles. The presence of long herbaceous rhizomes indicated serious isolation of the new species; hence, a new section Sulaimanicum is proposed to accommodate the new species. The new species is diploid with a chromosome number of 2n = 16. Detailed morphological description, illustrations, phylogenetic analyses based on sequences of plastid spacers (rpl32-trnL (UAG) and trnQ-rps16) and nuclear ITS, karyotype features, and a distribution map of the new species are provided.
Introduction
Allium L. (Amaryllidaceae J.St.-Hil.: Allioideae Herb.) is one of the largest monocot genera with currently more than 1,000 accepted species (Govaerts et al., 2021) mainly distributed throughout the Northern Hemisphere (Stearn, 1978; Stearn, 1992; Fritsch & Friesen, 2002; Friesen, 2022). The key centres of biodiversity are located in arid and sub-arid regions of Southwestern and Central Asia, occupying areas of the Mediterranean extending to the west of Asia Minor. Another considerably smaller centre is in western North America (Friesen et al., 2006; Nguyen et al., 2008; Li et al., 2010; Wheeler et al., 2013; Friesen, 2022). The genus is characterised by bulbs (often formed on rhizomes) enclosed in membranous, fibrous, or reticulate tunics, free or basally connate sepals, and usually a subgynobasic style (Friesen et al., 2006). The genus Allium is a member of the family Amaryllidaceae, subfamily Allioideae, and tribe Allieae (Chase et al., 2016; Friesen, 2022). Over 50 Allium species are widely grown globally or locally as cultivated plants. Also, very many wild species are collected by local people in nature as vegetables, for medicinal purposes, or as ornamental plants (Fritsch & Abbasi, 2013).
The enormous morphological, cytological, and genetic variabilities are reflected in the complicated taxonomic structure of the genus, consisting of 15 subgenera and 72 sections (Friesen et al., 2006), and more than 10 new sections were described during the last 10 years. All subsequent phylogenetic studies on the genus Allium have confirmed the three evolutionary lineages in the genus, including the monophyletic origin of all subgenera in the first and second evolutionary lineages (Li et al., 2010; Wheeler et al., 2013; Hauenschild et al., 2017; Xie et al., 2019; Costa et al., 2020; Xie et al., 2020; Friesen, 2022). The phylogenetic relationships in the third (youngest) lineage are less clear. According to the latest studies, many subgenera in the third lineage are not monophyletic. This mainly affects the subgenera Allium, Cepa (Mill.) Radić, Reticulatobulbosa (Kamelin) N. Friesen, Rhizirideum (G. Don ex Koch) Wendelbo, and Polyprason Radić (Li et al., 2016; Hauenschild et al., 2017; Xie et al., 2019; Friesen et al., 2020a; Xie et al., 2020; Friesen, 2022).
Forty-one Allium species were recorded in Flora of Pakistan (Nasir, 1975), with the description of Allium zhobicum N. Khan, A. Sultan & R. M. Fritsch and the addition of Allium caroli-henrici Wendelbo and Allium registanicum Wendelbo as new records for Pakistan (Khan et al., 2021); the genus representation in Pakistan was extended to 44 species.
During exploratory fieldwork in the Shinghar area (Sulaiman range) of Sherani district and Tor Ghar area of Musakhail District in northern Balochistan and the Sulaiman range in Dera Ismail Khan District of Khyber Pakhtunkhwa in 2019–2021 by Nazar Khan, Tahir Khan, and Kamran Ishaq, some Allium species were collected. Photos of two Allium species were sent to Reinhard Fritsch and Nikolai Friesen for identification. One of these was identified as Allium roylei Stearn, while the other plant, collected in the Sulaiman range, turned out to be an unfamiliar species. According to the morphological characters (inflorescences, flowers, and leaves), it was supposedly a new species from the Dagestanica section in the genus Allium, but the long herbaceous rhizome indicated serious isolation of this species. Leaf and seed samples were subsequently sent to Germany (Osnabrück) for molecular and cytological analyses. Molecular and cytological analyses have clearly shown the peculiarity of this species. In this work, we describe this plant as a new species, Allium sulaimanicum, and analyse its relationship and position in the Allium classification. With the addition of A. sulaimanicum, this genus is now represented by 45 species in Pakistan.
Material and methods
Taxon sampling
Plants of four accessions of A. sulaimanicum were collected in the Sulaiman Mountains during 2019–2021. DNA was isolated from silica gel-dried leaves. Newly sequenced accessions are marked with Am number in the trees, and their origin is shown in Table 1. To determine the position of the A. sulaimanicum in the genus, nuclear ITS sequences and rpl32-trnL (UAG) plastid fragments of representative species from most sections of the third evolutionary line were taken from NCBI GenBank (https://www.ncbi.nlm.nih.gov/nucleotide/). Some accessions from the first and second evolutionary lines were selected as outgroups (Friesen et al., 2006). Sequences from NCBI GenBank are marked with GenBank accession numbers on the trees. To find the closest relatives for A. sulaimanicum, we checked ITS, rpl32-trnL (UAG), and trnQ-rps16 sequences using the Nucleotide BLAST program of the NCBI GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
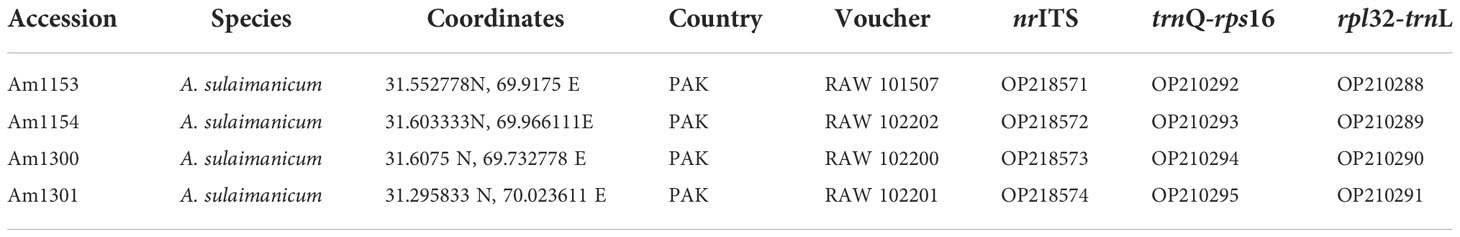
Table 1 Origin, source, and GenBank accession numbers of Allium sulaimanicum used for phylogenetic analyses.
DNA extraction, amplification, and sequencing
Total genomic DNA was isolated from leaves, dried in silica gel using the InnuPREPP Plant DNA Kit (Analytik Jena AG, Jena, Germany) according to the manufacturer’s instructions, and used directly in PCR amplification. The complete nuclear ribosomal ITS region (ITS1, 5.8S, and ITS2) was amplified using the primers ITS-A (Blattner, 1999) and ITS-4 (White et al., 1990). The PCR conditions for ITS followed those of Friesen et al. (2006). PCR conditions and primers for the chloroplast regions trnL-rpl32 and trnQ-rps16 were described in Shaw et al. (2007). PCR products were sent to Microsynth SeqLab (Göttingen, Germany; www.microsynth.seqlab.de) for sequencing. The sequences from all the individuals were manually edited in Chromas Lite 2.1 (Technelysium Pty Ltd., South Brisbane, QLD, Australia) and aligned with ClustalX (Thompson et al., 1997), and the alignment was manually corrected using MEGA 7 (Kumar et al., 2016).
Phylogenetic analyses
Both data sets (nrITS and the cpDNA trnL-rpl32 markers) were analysed separately for position identification in the third evolutionary lineage and to find the closest relatives of A. sulaimanicum through Fitch parsimony with the heuristic search option in PAUP version 4.0b10 (Swofford, 2002) with MULTREES, TBR branch swapping, and 100 replicates of random addition sequence. Gaps were treated as missing data. The consistency index (CI) (Kluge & Farris, 1969) was calculated to estimate the amount of homoplasy in the character set. The most parsimonious trees returned by the analysis were summarised in one consensus tree using the strict consensus method. Bootstrap analyses (bootstrap support (BS)) using 1,000 pseudoreplicates were performed to assess the support of the clades (Felsenstein, 1985). Bayesian phylogenetic analyses were also performed using MrBayes 3.1.23 (Ronquist & Huelsenbeck, 2003). The sequence evolution model was chosen following the Akaike information criterion (AIC) obtained from jModelTest2 (Dariba et al., 2012). Two independent analyses with four Markov chains were run for 10 million generations, sampling trees every 100 generations. The first 25% of trees were discarded as burn-in. The remaining 150,000 trees were combined into a single data set, and a majority-rule consensus tree was obtained along with posterior probabilities (PP). The alignments of both data sets can be found in Dryas: https://doi.org/10.5061/dryad.73n5tb313.
Cytology
Seeds were germinated in petri dishes on damp filter paper. The germinated seeds with root tips longer than 10 mm were kept on ice in distilled water overnight. They were then transferred to room temperature for 20 min and pre-treated for 3 h at room temperature in an aqueous solution of 0.1% colchicine. Roots were then fixed in a freshly prepared mixture of 96% ethanol and glacial acetic acid (3:1 v/v). Root tips were stained using hematoxylin according to the protocol reported by Smirnov (1968). Well-spread metaphase plates were electronically documented (digitally photographed), and finally, the chromosomes of the best plates were measured and pairwise arranged using the KaryoType software (Altinordu et al., 2016). Because the idiograms automatically assembled by the software were not satisfactory, we manually ordered the chromosome pairs according to their length and shape. The idiograms were designed using the bar graph function implemented in MS Excel®. The terminology of Levan et al. (1964) was applied. The karyotype was constructed using seven metaphase plates.
Results
Morphological description and distribution
A. sulaimanicum N. Khan, A. Sultan et N. Friesen sp. nov. Figures 1 and 2.
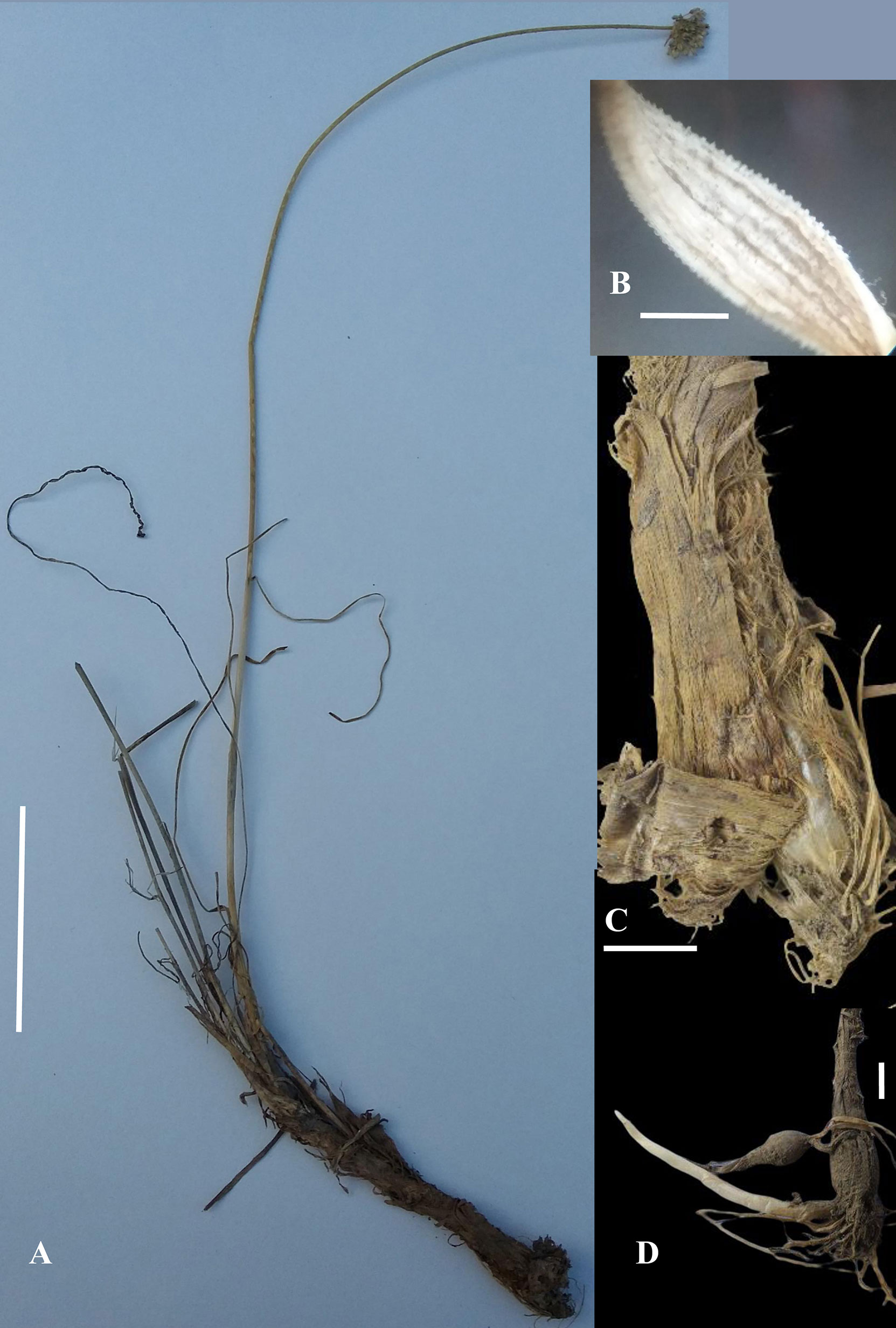
Figure 2 Allium sulaimanicum: (A) whole plant (scale bar: 8 cm) (B) Leaf in enlargement under a stereomicroscope (scale bar: c. 1 mm) (C) Bulb with outer coats brown coriaceous at the extreme base and otherwise fibrous with strips (scale bar: 6 mm) (D) Bulb with long rhizome (scale bar: 6 mm) (Photos by Nazar Khan except Fig. 2A by Amir Sultan and Fig. 2C by Muhammad Saleem).
Holotype: Khyber Pakhtunkhwa, Dera Ismail Khan District, Kaisa Ghar, Ahmadi Dirga, 31°33'10" N, 69°55'3" E, 2,022 m, N. K. Mandokhel, K. I. Bahadikhel & T. Khan, 16 July 2020 (RAW 101507).
Description
Bulb cylindrical, 4–9.5 cm long, 4–10 mm broad at the base, outer coats brown coriaceous at the extreme base and otherwise fibrous with strips, inner coats white membranous, in adult plants on long cylindrical rhizomes. Scapes 1–2, usually 1 with persistent old scapes, 25–45–(50) cm tall, covered at the base with scabrid leaf sheaths, cylindrical, bearing ridges and furrows, scabrid. Leaves linear 2–3, shorter than scape, 8–25 cm long, 1–3 mm broad, papillate along margins and veins, glabrous to sparsely pilose along veins with white hairs turning brown upon drying, veins 3–9. Spathe white with brownish veins and acuminate tips, split longitudinally unequally into two lobes. Umbel hemispherical, lax to densely flowered, pedicels unequal, 8–15 mm long, white to greenish at tips. Perigonium campanulate, tepals white to creamy white, 4.5–5 mm long, nerves brownish to purplish, ovate to elliptical, acute, recurved, inner tepals slightly broader at the base than outer. Stamens as long as to slightly longer than tepals, filaments connate at base and adnate to tepals, triangular at base, inner filaments slightly more dilated than outer at the base. Anthers yellow to brick red, 1–1.2 mm, basifixed. Ovary hexagonal, white, papillate/warty along angles, style 2–5 mm, stigma simple to slightly capitate sometime exserted. Seeds black, 1.5–2.5 × 0.8–1.2 mm, oblong to semi-orbicular, testa cells ± elliptical, periclinal walls ± even with 1–3 verrucae (Figure 2D).
Diagnosis
From the presumably related Allium maowenense J. M. Xu (NC Sichuan, China), it differs with open star-shaped flowers (closed in A. maowenense), stamens equal or only slightly longer than tepals (twice longer in A. maowenense), and cylindrical bulb (not ovoid). From Allium matiniae N. Friesen & M. Abbasi (NW Iran), it differs with stamens equal or only slightly longer than tepals (much shorter in A. matiniae) and base of inner filaments slightly more dilated than the outer ones (equal in A. matiniae). A. sulaimanicum differs from both species with long runner-like rhizomes.
Vernacular name
Da Khara Khokhai
Etymology
The epithet refers to the Sulaiman mountain range.
Distribution and conservation status
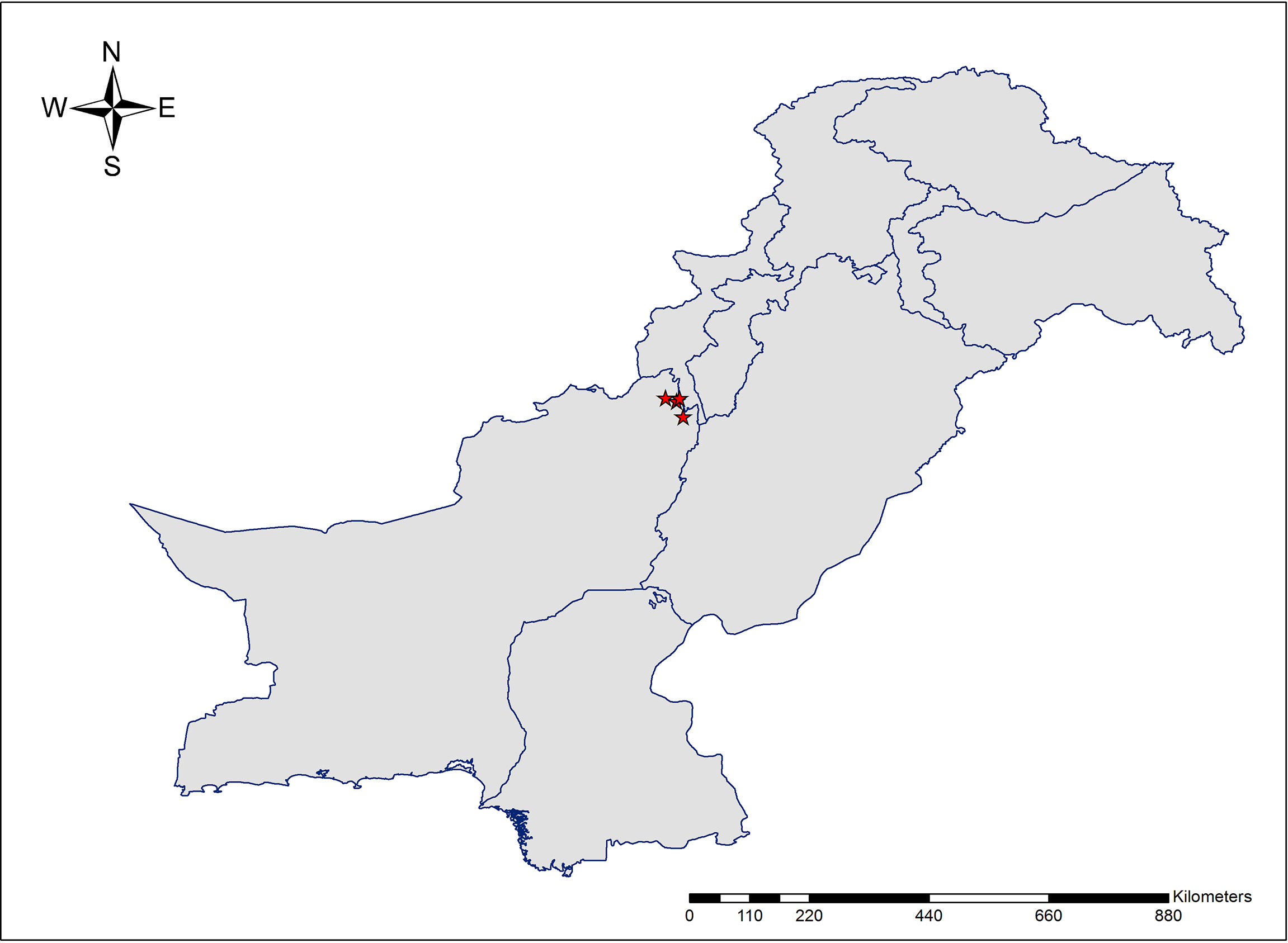
The new species is endemic to Pakistan and currently known from the Sulaiman range in southern parts of Dera Ismael Khan District and northern districts (Sherani and Musakhail) of Balochistan (Figure 3). Based on the current known distribution of A. sulaimanicum, its extent of occurrence is determined as 377.268 km2. However, further exploration in similar habitats in the Sulaiman range may yield further populations. Given the fire incidents in the Pinus gerardiana Wall. ex D.Don forest, it requires conservation measures.

Figure 3 Allium sulaimanicum. (A, B) Inflorescence (C) Flower section with tepals and stamens (scale bar: c 1.5 mm) (D) seed (scale bar: c 1 mm) (Photos by Nazar Khan).
Other specimens examined
Pakistan, Khyber Pakhtunkhwa, Dera Ismail Khan District, Takht-e-Sulaiman, 31 36 12 N, 69 57 58 E, 3000 m, N. K. Mandokhel, K. I. Bahadikhel & T. Khan, 16 July 2020 (RAW 102202); Pakistan, Balochistan, Sherani District, Shinghar, 31 36 27 N, 69 43 58 E, 2,800 m, N. K. Mandokhel & T. Khan, 15 September 2019 (RAW 102200); Musakhail District, Tor Ghar (Mizri Ghar), 31 17 45 N, 70 01 25 E, 2,366 m, N. K. Mandokhel, K. I. Bahadikhel & T. Khan, 19 September 2021 (RAW 102201).
Habitat and ecology
Found in shady places under chilgoza pine trees (P. gerardiana) or other shrubs and among herbaceous plants/grasses growing in gravelly loam to humus rich brownish to blackish soil at elevations of c. 2,000–3,000 m a.s.l.
Ethnobotany
Plants are not used by local people; flowers are grazed by goats.
Conservation status
The extent of occurrence: 377.268 km2
Allium subgen. Polyprasum Radić, Allium sect. Sulaimanicum N. Friesen sect. nov.
Typus: A. sulaimanicum N. Khan, A. Sultan et N. Friesen
Long runner-like cylindrical rhizomes of adult plants bear long-cylindrical bulbs with coriaceous tunics dissolving in strips and finally in reticulate fibres. Leaves linear. Perigonium campanulate, tepals white to creamy white, nerves brownish to purplish.
Phylogenetic analyses
Sequences of all four accessions of A. sulaimanicum (Am1153, Am1154, Am130, and Am1301) are identical in all sequenced plastid spacers (rpl32-trnL (UAG), trnQ-rps16, and trnL-trnF) and nuclear ITS.
Blast analysis
ITS
Blast analysis in the NCBI GenBank with ITS sequence of A. sulaimanicum accession Am1154 shows 86%–87% similarity with Allium species of Allium subgen. polyprason: HG794233 Allium cf. talassicum, 87.33%; FM945429 Allium hymenorhizum, 86.79%; HG794231 Allium kasteki, 86.79%; OM030262, OM030260 Allium palentinum, 86.64%; AJ250290 Allium carolinianum, 86.66%; HG794166 Allium tianschanicum, 87.18%; MF675014–MF675023 Allium forrestii, 86.57%.
rpl32-trnL (UAG)
The 10 best hits with rpl32-trnL spacer in the Blast analysis of NCBI GenBank are representatives of the Allium sect. Daghestanica: LR700256 A. matiniae, 95.41%; LR700253, LR700254 Allium daghestanicum, 95.40%; LR700250, LR700251, LR700252 Allium gunibicum, 95.40%; LR700255 A. daghestanicum, 95.28%; NC_042154 Allium chrysanthum, 93.76%; MH383268 Allium chrysocephalum, 93.65%; MH383267 Allium rude, 93.54%, followed in between with MK820610 Allium caeruleum, 93.34%, from Allium sect. Coerulea.
trnQ-rps16
Blast analysis in the NCBI GenBank with sequences of trnQ-rps16 of A. sulaimanicum shows very strong similarity with representatives of the Allium subgen. Cepa: MH159130 Allium altaicum and LT674586 Allium fistulosum, 98.51%; MT160180, NC_050981, MT300497, MT300496 Allium galanthum, 98.16%; KM088014, KF728079, Allium cepa 98.05%; MZ019479 Allium semenovii, 93.93% and LT699700, NC_057575, MN519208 Allium schoenoprasum, 97.48%.
Position of Allium sulaimanicum in the third evolutionary lineage
ITS analysis
The alignments of nrITS sequences (including the 5.8S gene) with 232 accessions of Allium species, comprising a selection of representatives from each subgenus and sections of the third evolutionary line, including four accessions of A. sulaimanicum, consist of 727 characters of which 510 variable characters are parsimony informative. Unweighted parsimony analysis of the 232 sequences resulted in about 14 million most parsimonious trees of 4,501 steps (CI = 0.2543). The substitution model TVM+G was chosen by AIC in JModeltest-2.1.7 for the Bayesian analysis. The parsimony and Bayesian analyses produce identical topology (see Figure S1). All A. sulaimanicum accessions form a monophyletic clade and stand alone in the third evolutionary line between representatives of the subgenera Polyprasum and Cepa (see Figure S1). The generalised ITS tree with sections and subgenera names is shown in Figure 4. Some subgenera in the third evolutionary lineage according to Friesen et al. (2006) are not monophyletic: this applies to A. subgenera Cepa, Reticulatobulbosa, Polyprason, and Allium. A. sulaimanicum accessions form a separate monophyletic clade inside the third evolutionary lineage: Bayesian PP = 1 and BS = 100 present strong support in the phylogenetic analyses.
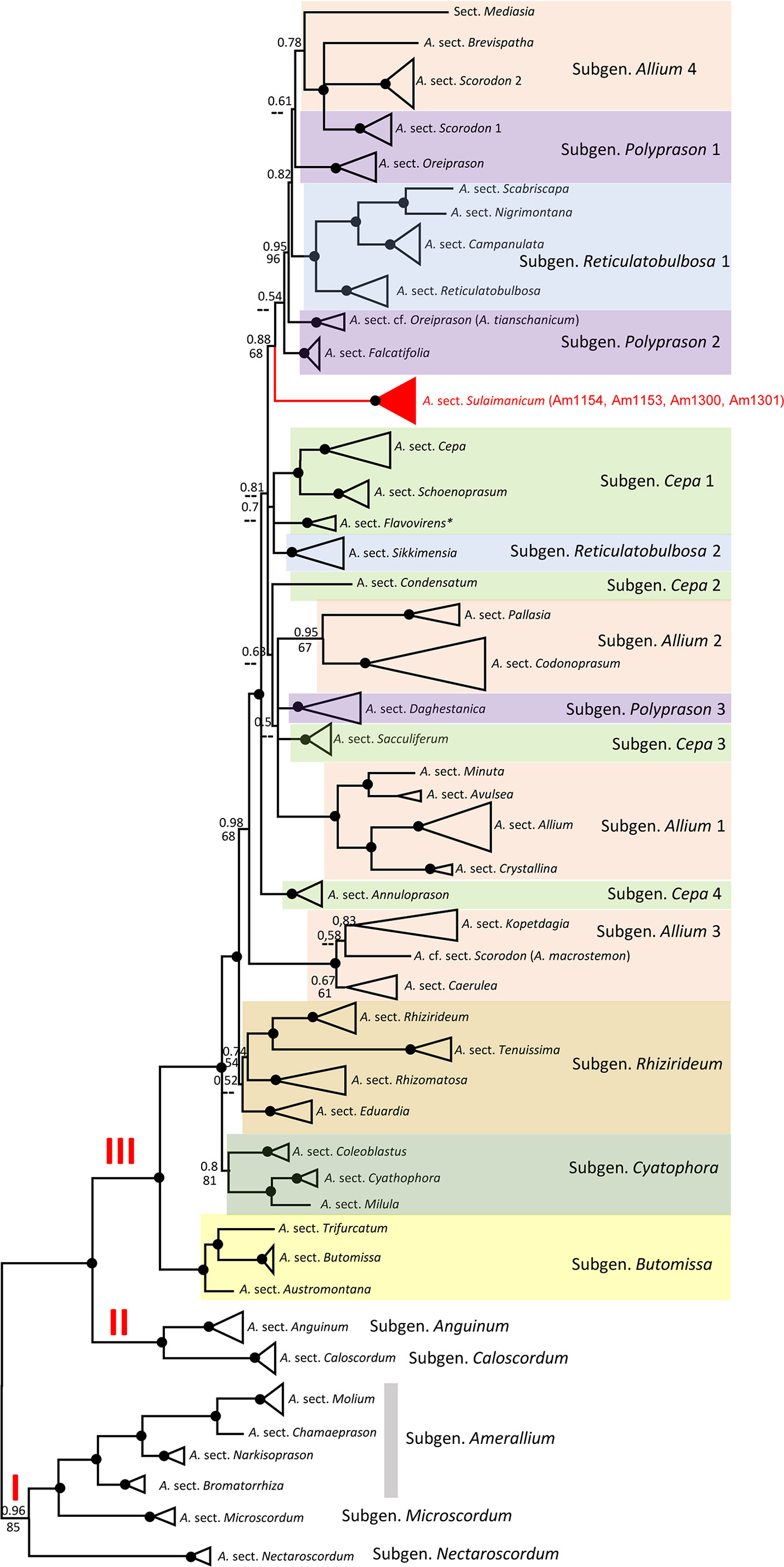
Figure 4 Generalised nrITS tree of the third evolutionary line of genus Allium. Numbers by nodes represent bootstrap support (1,000 replicates) and Bayesian probabilities. Roman numerals (I, II, and III) designate clades of three evolutionary lines. The joint presence of Bayesian probabilities over 0.98 and bootstrap support over 95% is indicated with a black dot.
CP analysis
The alignments of rpl32-trnL (UAG) sequences with 99 accessions of diverse Allium species (a selection of representatives from each subgenus and sections of the third evolutionary line, including four accessions of A. sulaimanicum) consist of 1,046 characters of which 282 variable characters are parsimony informative. Unweighted parsimony analysis of the 99 sequences resulted in about 1.541.841 most parsimonious trees of 841 steps (CI = 0.6385). The substitution model GTR+G was chosen by AIC in JModeltest-2.1.7 for the Bayesian analysis. The parsimony and Bayesian analyses produced identical topologies (see Figure S2).
The topology of the plastid tree, based only on an intergenic spacer (rpl32-trnL), is very different from the ITS tree. All four accessions of A. sulaimanicum were not resolved in the strongly supported clade (PP = 1; BS = 100) with Allium sect. Daghestanica and Allium subtilissimum Ledeb., whereas all species of Allium sect. Daghestanica formed a weakly supported clade (PP = 60; BS = 61) (see Figure S2). The generalised plastid tree with sections and subgeneric names is shown in Figure 5. The well-supported clade with representatives of the two Allium sections Falcatifolia and Coerulea form a sister clade to the clade with A. sulaimanicum but with very weak support (PP = 0.56 and no support in parsimony analysis). Furthermore, the A. sulaimanicum accessions belong to a strongly supported clade with representatives from the A. subgenera Reticulatobulbosa (only Allium sect. Campanulata) and Cepa. TrnQ-rps16 sequences are not known for all sections in the third evolutionary lineage and therefore could not be considered in the analysis.
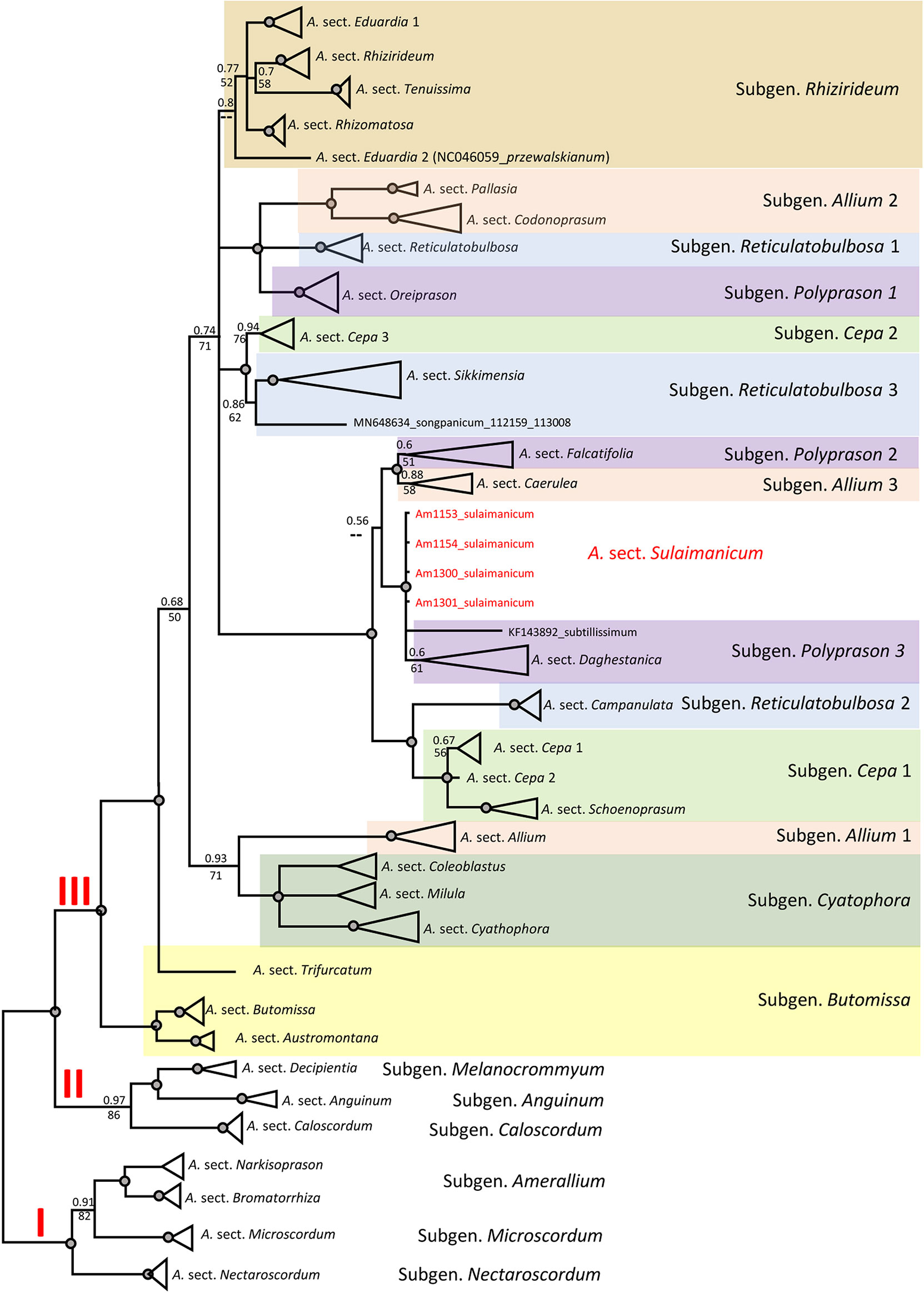
Figure 5 Generalised CP tree (rpl32-trnL (UAG) intergenic spacer) of the third evolutionary line of genus Allium. Numbers by nodes represent bootstrap support (1,000 replicates) and Bayesian probabilities. Roman numerals (I, II, and III) designate clades of three evolutionary lines. The joint presence of Bayesian probabilities over 0.98 and bootstrap support over 95% is indicated with a black dot.
Cytological analysis
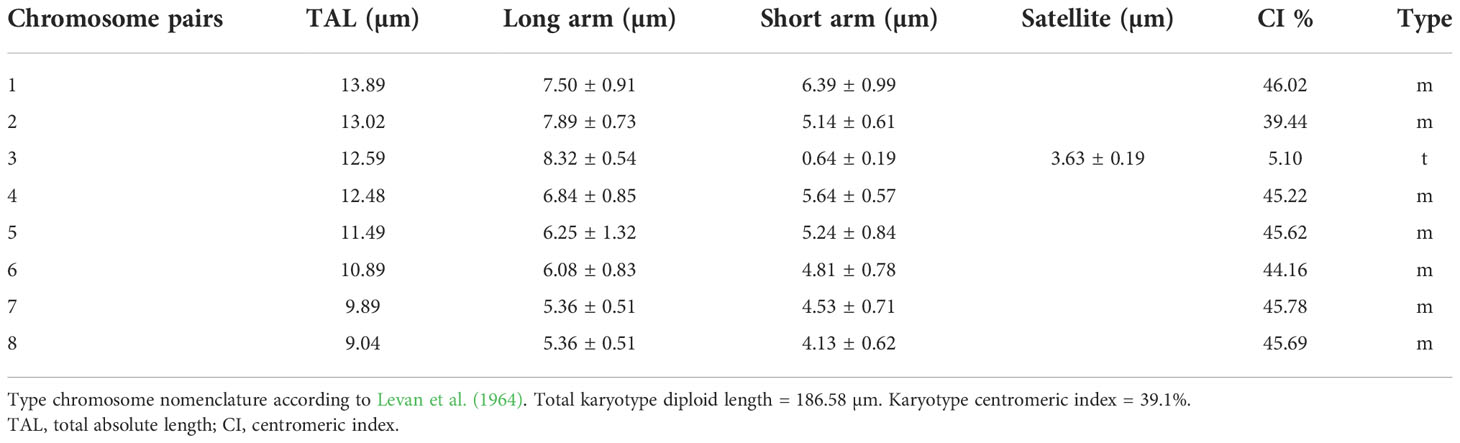
We examined the chromosome number and karyotype only from accession Am1301. The plant is diploid with 2n = 16. There are 14 metacentric chromosomes and two telocentric chromosomes with a big satellite (Table 2 and Figure 6).
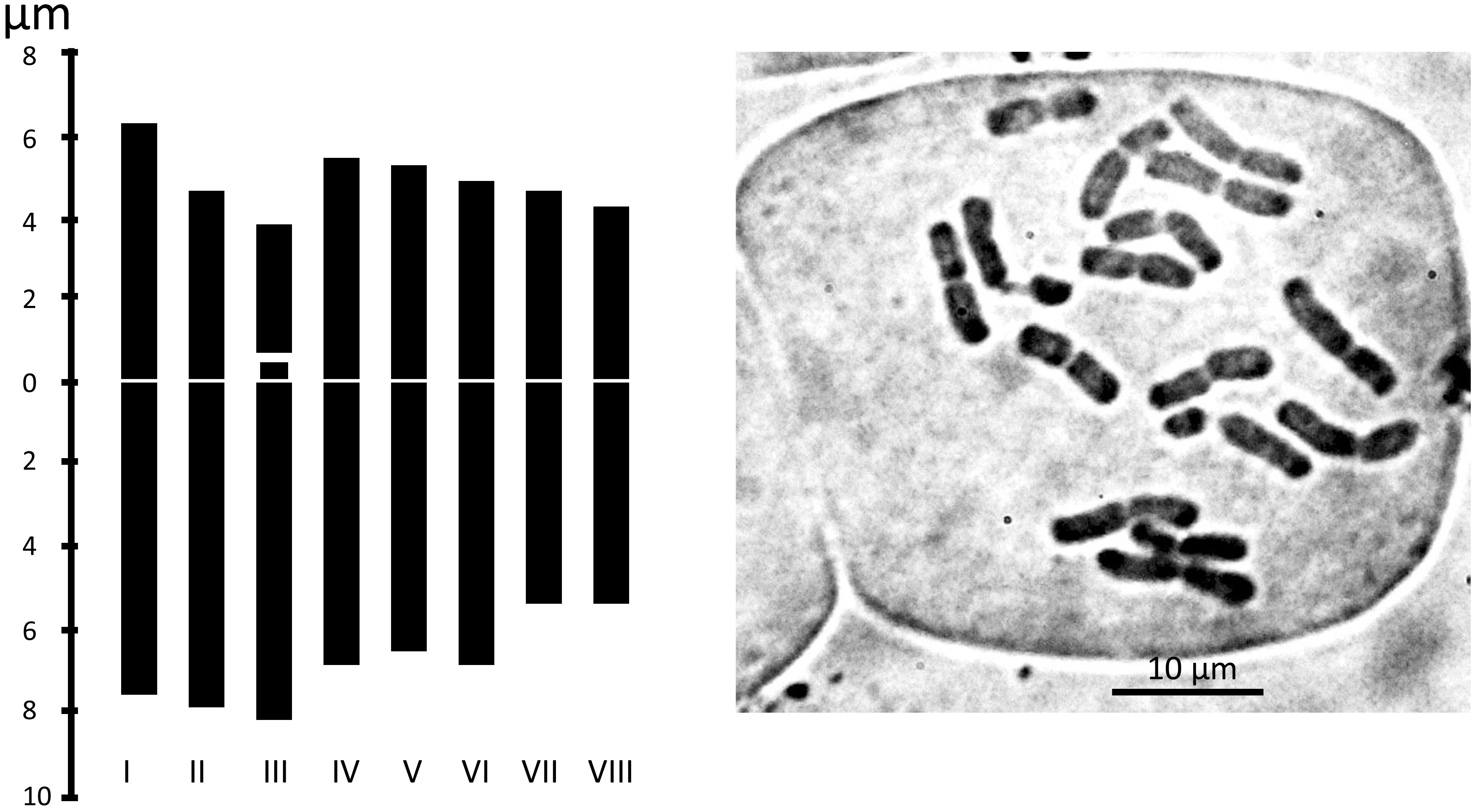
Figure 6 Metaphase plates and Idiograms of Allium sulaimanicum (Am1301). For the origin of the accession, see the Table 1.
Discussion
Some subgenera in the third evolutionary lineage according to Friesen et al. (2006) are not monophyletic in both nuclear (ITS) and plastid (rpl32-trnL) analyses, but the differences are not identical. This concerns the Allium subgenera Cepa, Reticulatobulbosa, Polyprason, and Allium. These results agree with previously published phylogenetic analyses (Li et al., 2016; Hauenschild et al., 2017; Xie et al., 2019; Friesen et al., 2020a; Xie et al., 2020; Friesen, 2022). The phylogenetic consequences for the non-monophyletic subgenera should be drawn from a detailed analysis in the future. The position of A. sulaimanicum inside the third evolutionary lineage is the most significant finding of the current study. In the ITS tree, the accessions of A. sulaimanicum form a separate monophyletic clade inside the third evolutionary lineage with strong support: Bayesian PPs = 1 and BS = 100 (see Figures 4, S1). In the plastid trees (Figures 5, S2), accessions of A. sulaimanicum form a strongly supported clade together with species of Allium sect. Daghestanica and A. subtilissimum. The sister group here is the clade with representatives of the Allium sect. Falcatifolia and Allium sect. Coerulea, which correlates well with blast analysis.
These discrepancies between nuclear (ITS) and plastid (rpl32-trnL) trees clearly point to a possible hybridogenic origin of A. sulaimanicum. One of the progenitors of A. sulaimanicum was certainly a representative of the Allium sect. Daghestanica, but the second parental taxon cannot be concluded from these data. Here, only the sequencing of the full genome can bring clarity. It should be noted that species of Allium sect. Daghestanica are distributed in three isolated areas: Europe, Caucasus, and China (Xie et al., 2019; Friesen et al., 2020b; Li et al., 2021). A. sulaimanicum is located in the large gap between Caucasian and Chinese localities. Another evidence of the relationship to some representatives of Allium sect. Daghestanica is the similarity of satellite chromosomes in A. sulaimanicum with satellite chromosomes in A. maowenense J. M. Xu (Xu et al., 1994), which also has telocentric chromosomes with very long satellites. It should be mentioned that other species in the section Daghestanica, in which the chromosomes were studied, have satellite chromosomes with very small satellites (A. chrysocephalum Regel and Allium herderianum Regel, Hongguan et al., 2005; A. gunibicum Miscz. ex Grossh., Zakirova and Vakhtina, 1974; Allium ericetorum Thore, Allium suaveolens Jacq., and Allium ochroleucum Walldst. & Kit., Májovský and Murín, 1985; Wetschnig, 1992).
Morphology of inflorescence and flowers shows some similarity with representatives of Allium sect. Daghestanica, the outer coats of bulbs are also similar, and only the long runner-like rhizome occurs rarely in the genus Allium (e.g., in Allium caespitosum Siev. ex Bong. & C.A.Mey from Allium sect. Rhizomatosa (Friesen et al., 2020a) and in some species in the section Allium (Mathew, 1996). The colour of the flowers is similar only in A. maowenense and A. matiniae from Allium sect. Daghestanica. All other representatives of Chinese species from Allium sect. Dagestanica have yellowish colour (see photos of all Chinese species of Allium sect. Daghestanica in Xie et al., 2019; Li et al., 2021). A. sulaimanicum was rarely detected hitherto, and we would like to encourage the scientific community to look for this new species whose morphological variation and ecological preferences remain insufficiently known yet.
Data availability statement
The data presented in the study are deposited in the NCBI GenBank, Accessions number OP218571 - OP218574, OP210288 - OP210295. Alignments are deposited in TataDryad doi: 10.5061/dryad.73n5tb313.
Author contributions
NK: Data curation, Formal analysis, Writing - review and editing. NF: Conceptualization, Formal analysis, Data curation, Writing - original draft, Writing - review and editing. AS: Resources, Data curation, Writing - review and editing. RF: Data curation, Formal analysis, Writing - review and editing., TK: Writing - review and editing. KI: Writing - review and editing. All authors contributed to the article and approved the submitted version.
Funding
Open access publication fee will be paid from the University of Osnabrück.
Acknowledgments
The authors are indebted to Mr. Amjad Khan for the distribution map.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1020440/full#supplementary-material
References
Altinordu, F., Peruzzi, L., Yu, Y., He, X. (2016). A tool for the analysis of chromosomes: KaryoType. Taxon 65, 586–592. doi: 10.12705/653.9
Blattner, F. R. (1999). Direct amplification of the entire ITS region from poorly preserved plant material using recombinant PCR. Biotechnology 27, 1180–1185. doi: 10.2144/99276st04
Chase, M. W., Christenhusz, M. J. M., Fay, M. F., Byng, J. W., Judd, W. S., Soltis, D. E., et al. (2016). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linnaean Soc. 181, 1–20. doi: 10.1111/boj.12385
Costa, L., Jimenez, H., Carvalho, R., Carvalho-Sobrinho, J., Escobar, I., Souza, G. (2020). Divide to conquer: Evolutionary history of Allioideae tribes (Amaryllidaceae) is linked to distinct trends of karyotype evolution. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00320
Dariba, D., Taboada, G. L., Doallo, R., Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Friesen, N. (2022). “Chapter 1. Genus Allium: Evolution, classification, and domestication,” in Rabinowitch, H.D., Brian, t. (Eds.) Edible Alliums: Modern biology, production and uses (Wallingford, UK: CABI Org.; in press).
Friesen, N., Abbasi, M., Murtazaliev, R., Fritsch, R. M. (2020b). Allium matinae – a new species from northwestern Iran. Phytotaxa 433.3, 181–189. doi: 10.11646/phytotaxa.433.3.1
Friesen, N., Fritsch, R. M., Blattner, F. R. (2006). Phylogeny and new intrageneric classification of Allium l. (Alliaceae) based on nuclear rDNA ITS sequences. Aliso 22, 372–395. doi: 10.5642/aliso.20062201.31
Friesen, N., Smirnov, S., Herden, T., Oyuntsetseg, B., Shmakov, A., Hurka, H. (2020a). Allium species of section Rhizomatosa, early members of the Central Asian steppe vegetation. Flora 263, 151536. doi: 10.1016/j.flora.2019.151536
Fritsch, R. M., Abbasi, M. (2013). A taxonomic review of Allium subg. Melanocrommyum in Iran (Germany: Leibniz Institut für Pflanzengenetik und Kulturpflanzenforschung), 240pp. Available at: http://pgrc.ipk-gatersleben.de/allium/.
Fritsch, R. M., Friesen, N. (2002). “Evolution, domestication and taxonomy,” in Rabinowitch HD, Currah L. (Eds) Allium Crop Science: Recent Advances (Wallingford: CABI Publishing), 5–30. doi: 10.1079/9780851995106.0005
Govaerts, R., Kington, S., Friesen, N., Fritsch, R., Snijman, D. A., Marcucci, R., et al. (2021) World checklist of Amaryllidaceae. Facilitated by the Royal Botanic Gardens, Kew. Available at: http://apps.kew.org/wcsp/ (Accessed 20.03.2022).
Hauenschild, F., Favre, A., Schnitzler, J., Michalak, I., Freiberg, M., Muellner-Riehl, A. N. (2017). Spatio-temporal evolution of Allium L. in the Qinghai–Tibet–Plateau region: Immigration and in situ radiation. Plant Divers. 39, 167–179. doi: 10.1016/j.pld.2017.05.010
Hongguan, T., Lihua, M., Shiqimg, A., Jianquaan, L. (2005). Origin of the qinghai-Tibetan plateau endemic Milula (Liliaceae): further insights from karyological comparisons with Allium. Caryologia 58 (4), 320–331. doi: 10.1080/00087114.2005.10589470
Khan, N., Fritsch, R. M., Sultan, A., Khan, T. (2021). Allium (Amaryllidaceae) species in Pakistan: two new records and a new species from zhob (Balochistan). Pak. J. Bot. 53 (5), 1759–1765. doi: 10.30848/PJB2021-5(1
Kluge, A. G., Farris, J. S. (1969). Quantitative phyletics and the evolution of anurans. Syst. Zool. 18, l–32. doi: 10.1093/sysbio/18.1.1
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Levan, A., Fredga, K., Sandberg, A. A. (1964). Nomenclature for centromeric position on chromosomes. Hereditas 52, 201–220. doi: 10.1111/j.1601-5223.1964.tb01953.x
Li, M.-J., Guo, X.-L., Yu, H.-X., He, X.-J. (2021). Out of the Qinghai–Tibetan plateau and rapid radiation across Eurasia for Allium section Daghestanica (Amaryllidaceae). AoB Plants 13, 3. doi: 10.1093/aobpla/plab017
Li, Q. Q., Zhou, S. D., He, X.-J., Yu, Y., Zhang, Y. C., Wei, X. Q. (2010). Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China. Ann. Bot. 106 (5), 709–733. doi: 10.1093/aob/mcq177
Li, Q. Q., Zhou, S. D., Huang, D. Q., He, X.-J., Wei, X. Q. (2016). Molecular phylogeny, divergence time estimates and historical biogeography within one of the world’s largest monocot genera. AoB Plants 8, plw041. doi: 10.1093/aobpla/plw041
Májovský, J., Murín, A. (1985). Karyotaxonomisches studium des Allium ericetorum aggr. Acta Fac. Rerum Nat. Univ. Comenianae Bot. 32, 3–20.
Nasir, E. (1975). “Alliaceae,” in Nasir, E., Ali, S. I. (Eds.), Flora of West Pakistan. No. 83. Stewart Herbarium (Rawalpindi, Pakistan: Gordon College), 1–31.
Nguyen, N. H., Driscoll, H. E., Specht, C. D. (2008). A molecular phylogeny of the wild onions (Allium; Alliaceae) with a focus on the western north American center of diversity. Mol. Phylogenet. Evol. 47 (3), 1157–1172. doi: 10.1016/j.ympev.2007.12.006
Ronquist, R., Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Shaw, J. E. B., Lickey, E. E., Schilling, E. E., Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 94, 275–288. doi: 10.3732/ajb.94.3.275
Smirnov, Y. A. (1968). Accelerated method of study of somatic chromosomes of fruit plants. Tsitol. (Moskva) 10, 1601–1602.
Stearn, W. T. (1978). European Species of Allium and allied genera of alliaceae: a synonymic enumeration. Annales Musei Goulandris 4, 83–198.
Stearn, W. T. (1992). How many species of Allium are known? Kew Magazine 9, 180–182. doi: 10.1111/j.1467-8748.1992.tb00096
Swofford, D.–L. (2002). PAUP*: Phylogenetic analysis using parsimony (* and other methods). version 4 (Sunderland, Massachusetts: Sinauer Associates).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The clustal X window interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Wetschnig, W. (1992). Chromosomenzahlen kärntner gefässpflanzen (Teil 3): Karyologie und Verbreitung der Allium-Arten (Alliaceae) in Kärnten. Carinthia II 182, 497–533.
Wheeler, E. J., Mashayekhi, S., McNeal, D. W., Columbus, J. T., Pires, J. C. (2013). Molecular systematics of Allium subgenus Amerallium (Amaryllidaceae) in north America. Am. J. Bot. 100, 701–711. doi: 10.3732/ajb.1200641
White, T. J., Bruns, T., Lee, S., Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: a guide to methods and applications (New York: Academic Press), 315–322. doi: 10.1007/BF00224530
Xie, D. F., Tan, J. B., Yu, Y., Gui, L. J., Su, D. M., Zhou, S. D., et al. (2020). Insights into phylogeny, age and evolution of Allium (Amaryllidaceae) based on the whole plastome sequences. Ann. Bot. 125, 1039–1055. doi: 10.1093/aob/mcaa024
Xie, D. F., Yu, H. X., Price, M., Xie, C., Deng, Y. Q., Chen, J. P., et al. (2019). Phylogeny of Chinese Allium species in section Daghestanica and adaptive evolution of Allium (Amaryllidaceae, Allioideae) species revealed by the chloroplast complete genome. Front. Plant Sci. 10, e460. doi: 10.3389/fpls.2019.00460
Xu, J. M., Xue, P. F., Zhu, S. M., Jing, W. C. (1994). A new species of Allium l. Sichuan Karyotype. Acta Phytotax. Sin. 32 (4), 356–358.
Zakirova, R. O., Vakhtina, L. I. (1974). “Cytophotometrical investigation of some species of the genus Allium section Cepa Prokh. and Rhizirideum Don,” in Funktsional’naja morfologija, genetika i biokhimija kletki. (Functional morphology, genetics and biochemistry of cell), vol. 16. (Russian: Leningrad vyp), 67–70.
Keywords: Sulaiman range, ITS, rpl32-trnL, phylogeny, chromosome
Citation: Khan N, Friesen N, Sultan A, Fritsch RM, Khan T and Ishaq K (2022) Allium sulaimanicum: A new Allium species and section from Pakistan. Front. Plant Sci. 13:1020440. doi: 10.3389/fpls.2022.1020440
Received: 16 August 2022; Accepted: 10 November 2022;
Published: 14 December 2022.
Edited by:
Paulo Takeo Sano, Departamento de Botânica, Universidade de São Paulo, BrazilReviewed by:
Zhechen Qi, Zhejiang Sci-Tech University, ChinaLlorenç Saez, Universitat Autònoma de Barcelona, Spain
Copyright © 2022 Khan, Friesen, Sultan, Fritsch, Khan and Ishaq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nikolai Friesen, nfriesen@uni-osnabrueck.de
 Nazar Khan
Nazar Khan Nikolai Friesen
Nikolai Friesen Amir Sultan3
Amir Sultan3