- 1Department of Plant Sciences, University of California, Davis, Davis, CA, United States
- 2College of Agronomy, Northwest A&F University, Yangling, China
- 3School of Plant Sciences and Food Security, Tel Aviv University, Tel Aviv, Israel
Meiotic pairing between homoeologous chromosomes in polyploid wheat is inhibited by the Ph1 locus on the long arm of chromosome 5 in the B genome. Aegilops speltoides (genomes SS), the closest relative of the progenitor of the wheat B genome, is polymorphic for genetic suppression of Ph1. Using this polymorphism, two major suppressor loci, Su1-Ph1 and Su2-Ph1, have been mapped in Ae. speltoides. Su1-Ph1 is located in the distal, high-recombination region of the long arm of the Ae. speltoides chromosome 3S. Its location and tight linkage to marker Xpsr1205-3S makes Su1-Ph1 a suitable target for introgression into wheat. Here, Xpsr1205-3S was introgressed into hexaploid bread wheat cv. Chinese Spring (CS) and from there into tetraploid durum wheat cv. Langdon (LDN). Sequential fluorescence in situ hybridization and genomic in situ hybridization showed that an Ae. speltoides segment with Xpsr1205-3S replaced the distal end of the long arm of chromosome 3A. In the CS genetic background, the chromosome induced homoeologous chromosome pairing in interspecific hybrids with Ae. peregrina but not in progenies from crosses involving alien disomic substitution lines. In the LDN genetic background, the chromosome induced homoeologous chromosome pairing in both interspecific hybrids and progenies from crosses involving alien disomic substitution lines. We conclude that the recombined chromosome harbors Su1-Ph1 but its expression requires expression of complementary gene that is present in LDN but absent in CS. We suggest that it is unlikely that Su1-Ph1 and ZIP4-1, a paralog of Ph1 located on wheat chromosomes 3A and 3B and Ae. tauschii chromosome 3D, are equivalent. The utility of Su1-Ph1 for induction of recombination between homoeologous chromosomes in wheat is illustrated.
Introduction
In most allopolyploid plants, only homologous chromosomes pair in meiosis and only bivalents are present at metaphase I (MI); pairing between homoeologous chromosomes (heterogenetic chromosome pairing) is excluded (Jenczewski and Alix, 2004). The best-known example of genetic exclusion of heterogenetic chromosome pairing is in tetraploid and hexaploid wheat (Okamoto, 1957; Riley and Chapman, 1958; Sears and Okamoto, 1958).
The chromosome complement of tetraploid wheat (Triticum turgidum L., 2n = 4x = 28) consists of the A genome, which was contributed by T. urartu Thum., and the B genome, which was contributed by an extinct or undiscovered species closely related to Aegilops speltoides Tausch (genomes SS≈BB) (Dvorak and Zhang, 1990; Dvorak et al., 1993). The chromosome complement of hexaploid wheat (T. aestivum L., 2n = 6x = 42) consists of the A and B genomes of tetraploid wheat and the D genome, which was contributed by Ae. tauschii Coss. (Kihara, 1944; McFadden and Sears, 1946; Wang et al., 2013). While the chromosomes of these diploid species extensively pair in hybrids among them, virtually no chromosome pairing take places in haploids derived from polyploid wheat (Kimber and Riley, 1963; McGuire and Dvořák, 1982; Jauhar et al., 1991, 1999). This paradox is caused by the expression of the Ph1 locus on chromosome 5B of tetraploid and hexaploid wheat, which prevents pairing between homoeologous chromosomes (Okamoto, 1957; Riley and Chapman, 1958; Sears and Okamoto, 1958). If Ph1 is absent due to aneuploidy (Riley, 1960) or a deletion, such as ph1b in hexaploid wheat (Sears, 1977) and ph1c in tetraploid wheat (Giorgi, 1978), meiotic pairing of homoeologous chromosomes is restored.
The Ph1 locus on chromosome 5B was initially associated with a cluster of cyclin-dependent kinase 2 (Cdk2)-like protein genes including a DNA fragment translocated from chromosome 3A (Griffiths et al., 2006). The translocated fragment was shown to contain TaZIP4-B2, a highly expressed paralog of TaZIP4-1, which is located on wheat chromosomes 3A (TaZIP4-A1), 3B (TaZIP4-B1), and 3D (TaZIP4-D1) (Rey et al., 2017).
Genes suppressing Ph1 and promoting homoeologous chromosome pairing have been reported in several wheat relatives. Examples are chromosome 5U of Ae. umbellulata Zhuk. (Riley et al., 1973), chromosomes 3E, 4E, and 5E of Lophopyrum elongatum (Host) Á. Löve (Dvořák, 1987), and chromosome 5Mg of Ae. geniculata Roth (Koo et al., 2016). Polymorphism for the suppression of Ph1 was observed in Ae. speltoides and Amblyopyrum muticum (Boiss.) Eig (Dover and Riley, 1972; Dvořák, 1972; Kimber and Athwal, 1972). Using this polymorphism, major suppressors of Ph1 were mapped on Ae. speltoides chromosome arms 3SL (Su1-Ph1) and 7SL (Su2-Ph1) (Dvorak et al., 2006b).
The first use of Ph1 suppression to introgress a gene from a wheat relative into wheat employed hybridization of a wheat cytogenetic stock with Ae. speltoides (Riley et al., 1973). The presence of the Ae. speltoides genome induced recombination between the alien and wheat homoeologs. A logical extension of this idea is to introgress one of the Ae. speltoides suppressors into wheat. Such a wheat genetic stock would greatly simplify introgression of alien genes into wheat. Induction of recombination between homoeologous chromosomes would require nothing more than substituting the alien chromosome for a wheat homoeolog and crossing the substitution line with the Ph1 suppressor line. The F1 would be doubly monosomic for the homoeologous chromosomes targeted for recombination and the expression of Ph1 would be simultaneously suppressed by heterozygosity for the Ph1 suppressor, provided that the suppressor is dominant. The first attempt to produce such a genetic stock resulted in the introgression of an Ae. speltoides suppressor named Ph1I (Chen et al., 1994). The suppression of Ph1 in this line is relatively weak (Chen et al., 1994). It is also not known what Ae. speltoides gene was introgressed and where Ph1I is located in the wheat genome (Li et al., 2011). This situation has precluded the use of marker assisted selection (MAS) in genetic manipulations with this stock.
Another introgression of an Ae. speltoides suppressor into wheat occurred inadvertently as a by product of introgression of the leaf rust resistance gene Lr66 located on chromosome 3S in Ae. speltoides (Marais et al., 2010). The introgressed gene could have been Su1-Ph1. The introgression was accompanied by sterility and has not been practically exploited.
Here, we report introgression of Su1-Ph1 into the genetic background of hexaploid wheat (T. aestivum) cv. Chinese Spring (CS) and from there into the genetic background of tetraploid durum wheat (T. turgidum ssp. durum) cv. Langdon (LDN) utilizing MAS with Xpsr1205-3S. The marker is 0.2 to 0.4 cM distal to Su1-Ph1. MAS was aided by the development of an Ae. speltoides-specific assay for the Xpsr1205-3S haplotype (Dvorak et al., 2006b). The expression of the introgressed Su1-Ph1 in the CS and LDN genetic backgrounds is evaluated and used to test the hypothesis that MAS for Xpsr1205-3S resulted in the introgression of Su1-Ph1. The locations of the Su1-Ph1 relative to the ZIP4-1 loci in the genomic sequences of chromosomes 3A and 3B of wild emmer (Avni et al., 2017) and that of chromosome 3D of Ae. tauschii (Luo et al., 2017) are used to test the hypothesis that Su1-Ph1 is equivalent to ZIP4-1. A strategy for using the introgressed Su1-Ph1 for introgression of alien genes in wheat is suggested.
Materials and Methods
Genetic Stocks
The initial material for introgression of Su1-Ph1 into CS and LDN was Ae. speltoides F4 family #134. The family was derived from an F2 plant #134 in a population used for Ph1 suppressor mapping (Dvorak et al., 2006b). The plant was homozygous for an active allele at Su1-Ph1 and an inactive allele at Su2-Ph1. Family #134 inherited chromosome 3S with Su1-Ph1 from Ae. speltoides accession PI 369609. The family was crossed with CS (accession DV148) to produce a CS × Ae. speltoides hybrid. To produce an octoploid amphiploid (2n = 8x = 56), the CS × Ae. speltoides #134 F1 hybrid was removed from its pot and the crown was immersed into 0.6% aqueous solution of colchicine overnight. The disomic substitution (DS) line of L. elongatum chromosome 3E for CS chromosome 3B, designated as DS3E(CS3B) (Tuleen and Hart, 1988) was employed in the initial stages of Su1-Ph1 intorgression into CS. In addition, the DS lines of Ae. searsii chromosomes 1Sse, 5Sse, and 6Sse for their CS homoeologs, designated DS1Sse(CS1A), DS1Sse(CS1B), DS1Sse(CS1D), DS5Sse(CS5A), DS6Sse(CS6A), and DS6Sse(CS6D) (Friebe et al., 1995) were provided by N. E. Tuleen, Texas A&M University, College Station, TX, United States. The DS line of L. elongatum chromosome 1E in LDN, DS1E(LDN1A) and DS1E(LDN1B) (Jauhar and Peterson, 2011) were supplied by P. Jauhar, University of North Dakota, Fargo.
Molecular Markers
A genome-specific PCR assay for Xpsr1205-3S (Dvorak et al., 2006b) was used in MAS for Su1-Ph1 during backcrossing in the CS and LDN genetic backgrounds. Sequence information for wheat ESTs BE426080 and CD454867 and conserved primers of BE426080 (Forward- TGCACTTGCAAATCAAAAGC; Reverse- CGATCTTGCCACTCTTCTCC) and CD454867 (Forward- AGCTCCAGCAATCCTCTCAA; Reverse- GATGGTCGGCTATGCTCTTC) were obtained from the wheat single- nucleotide polymorphism (SNP) database1 (Akhunov et al., 2010). Using these conserved primers, amplicons were amplified from genomic DNAs of Ae. searsii accession TE10, DS5Sse(CS5B), CS, and LDN and were treated with ExoSAP-IT (USB) and Sanger sequenced according to the manufacturers protocol (Applied Biosystems, Foster City, CA, United States). A homology search was then performed using BLAST at the bread wheat chromosome-based survey sequence2 site with these sequences. Alignments and comparisons of these sequences were used to discover SNPs at which the Ae. searsii nucleotide sequence differed from the wheat A-, B-, and D-genome nucleotide sequences. These polymorphisms were used to design Ae. searsii genome-specific primers. Their specificity was tested by PCR of DNAs of Ae. searsii accession TE10, DS5Sse(CS5B), CS, and LDN. Two Ae. searsii chromosome 5Sse-specific SNP markers, BE426080-5Sse (Forward- TTCTAGTAGAAGCTATTTCATGAGTAACTG; Reverse- CGATCTTGCCACTCTTCTCC) and CD454867-5Sse (Forward- AGCTCCAGCAATCCTCTCAA; Reverse- GAAAGGAGTTCAATGTGCTTCG) were developed and used for selection of plants with the 5Sse chromosome and for study of recombination between 5Sse and 5B.
Markers for studying recombination between LDN chromosome 1A and L. elongatum chromosome 1E were developed as follows. Markers were selected based upon their position on the Ae. tauschii SNP genetic map (Luo et al., 2013). Conserved primers were designed as described previously (Akhunov et al., 2010) and used to generate amplicons from Ae. tauschii accession AL8/78, Ae. speltoides line 134, T. urartu accession G1812, and L. elongatum. The amplicons were sequenced according to the protocol above. Amplicon sequences were aligned and differences between the sequences leveraged to design genome specific primers. E-genome specific primers (Supplementary Table S1) were tested for PCR amplification in genomic DNA of L. elongatum, T. aestivum cv. Chinese Spring, and DS1E(LDN1A) and DS1E(LDN1B).
The PCR reaction contained 1 unit of Taq DNA polymerase, 3 mM MgCl2, 50 pmol of forward and reverse primers, and 50 ng of template. The reaction conditions were as follows. DNA was denatured at 98°C for 5 min, which was followed by 40 cycles consisting of 96°C for 30 s, 30 s at annealing temperature, and extension at 72°C for 2 min. The 40 cycles were terminated by maintaining 72°C for 5 min. The amplicons were visualized by electrophoresis in a 2% agarose gel and stained with ethidium bromide.
Cytological Observations
To verify that Su1-Ph1 was introgressed into CS, BC4F2 plants heterozygous for Xpsr1205-3S were crossed with accessions G634, G637, G666, G1326, and G4984 of Ae. peregrina (Hack. in J. Fraser) Maire & Weiller (2n = 4x = 28) provided originally under the synonym Ae. variabilis by B. L. Johnson, University of California, Riverside, CA, United States. The presence of Xpsr1205-3S and the level of chromosome pairing was determined in each hybrid. Ae. peregrina accessions G1326 and G666 were also crossed with CS and the ph1b deletion mutant (Sears, 1977), respectively. To determine whether Su1-Ph1 was active in the CS genetic background, B5F2 introgression plants heterozygous for Xpsr1205-3S were crossed with DS lines for Ae. searsii chromosomes 1Sse and 6Sse (Friebe et al., 1995) (provided by N. E. Tuleen, Texas A&M University, College Station, TX, United States). Hybrids were genotyped with Xpsr1205-3S and chromosome pairing was examined.
For chromosome pairing studies, immature spikes were collected from greenhouse-grown plants, fixed in freshly prepared Carnoy’s 6:3:1 (ethanol/acetic-acid/chloroform) fixative for 24 h at room temperature, and then stored in 70% ethanol. Squashes of pollen mother cells (PMCs) were stained with 2% acetocarmine. Meiotic chromosome pairing was scored in 20–30 meiocytes per plant. In the meiotic configurations, univalents, bivalents, trivalents, quadrivalents, and quinquevalents are indicated by I, II, III, IV, and V, respectively. The number of such configurations are indicated by an Arabic numeral preceding the symbol.
Mitotic chromosome spreads of tetraploid introgression lines were subjected to sequential fluorescence in situ hybridization (FISH) and genomic in situ hybridization (GISH). Chromosome spread preparation, probe labeling, and in situ hybridization were carried out according to the methods previously described by Li et al. (2014). In GISH procedures, genomic DNA of Ae. speltoides PI 369609 was labeled with ChromaTide Alexa Fluor-488-5-dUTP (Thermo Fisher Scientific, Waltham, MA, United States) as probe, and genomic DNA of LDN was used as blocking DNA. In FISH procedures, oligonucleotide probes Oligo-pTa535 and Oligo-pSc119.2 were used. Oligo-pTa535 and Oligo-pSc119.2 were reported to be equivalent to repetitive sequence probes in distinguishing wheat A-, B-, and D-genome chromosomes (Tang et al., 2014). Oligonucleotide probes labeled with 6-carboxyfluorescein (6-FAM) or 6- carboxytetramethylrhodamine (Tamra) at the 5′ end, were synthesized by Thermo Fisher Scientific. Photographs were captured with cellSens Standard 1.8 software (Olympus Corporation, Tokyo, Japan) on Olympus BX53 fluorescence microscope with a DP80 microscope digital camera (Olympus Corporation, Tokyo, Japan), and then processed with Adobe Photoshop CS 6.0 (Adobe Systems Incorporated, San Jose, CA, United States).
Results
Introgression of the Ae. speltoides Ph1 Suppressor (Su1-Ph1) into Hexaploid and Tetraploid Wheat
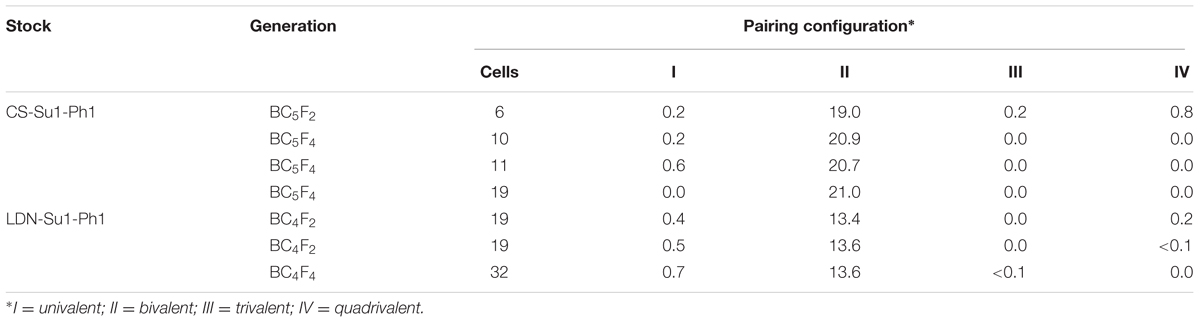
In our effort to introgress the entire Ae. speltoides chromosome 3S into wheat, the 56-chromosome amphiploid CS × Ae. speltoides was crossed with DS3E(CS3B) (Figure 1A). Ten F1 plants positive for the Ae. speltoides Xpsr1205-3S haplotype were obtained. The plants were male-sterile and were backcrossed once as females to DS3E(CS3B). The BC1F1 plants were again male sterile and were backcrossed as females to CS. Five male-sterile BC2F1 plants positive for Xpsr1205-3S were obtained and backcrossed once more as the female and male-fertile progeny was three times backcrossed as males to CS. Plants bearing Xpsr1205-3S were selected in each generation. The family of hexaploid introgression lines with Xpsr1205-3S in the CS genetic background was designated as CS-Su1-Ph1.
FIGURE 1. Introgression of Su1-Ph1 into hexaploid and tetraploid wheat. (A) Breeding scheme culminating in the development of CS-Su1-Ph1 and LDN-Su1-Ph1. Xpsr1205-3S is an Ae. speltoides-specific marker tightly linked to Su1-Ph1. (B) Spike and seeds of T. aestivum cv. Chinese Spring (CS) and a CS-Su1-Ph1 introgression line. (C) Spike and seeds of T. turgidum ssp. durum cv. Langdon (LDN) and LDN-Su1-Ph1.
To introgress Su1-Ph1 from CS-Su1-Ph1 into T. turgidum ssp. durum cv. LDN, a heterozygous BC5F2 plant (37∗95∗4∗14-8) was crossed with LDN and six Xpsr1205-3S-positive F1 plants were grown. The pentaploid hybrid was male sterile. It was backcrossed as the female parent to LDN four times, always selecting for the Xpsr1205-3S haplotype in progeny. Fertile plants positive for Xpsr1205-3S were ultimately obtained. They were designated as LDN-Su1-Ph1.
The CS-Su1-Ph1 lines were similar to CS in their morphology except for a pyramidal spike shape (Figure 1B). All CS-Su1-Ph1 plants, irrespective of the presence or absence of Xpsr1205-3S, had significantly lower seed set than CS but there was no difference in seed set between siblings with and without Xpsr1205-3S (Table 1) (one-way ANOVA, α = 0.05, LSD). Except for a single plant with a translocation, CS-Su1-Ph1 plants did not show multivalent chromosome pairing (Table 2).
TABLE 1. Mean number of seeds per spikelet in CS-Su1-Ph1 and LDN-Su1-Ph1 plants with and without Xpsr1205-3S, Chinese Spring and Langdon.
LDN-Su1-Ph1 lines were similar to LDN in spike shape, seed shape, and seed size. LDN-Su1-Ph1 seeds had darker pericarp than those of LDN (Figure 1C). LDN-Su1-Ph1 were less fertile than LDN (P = 0.001, t-test, N = 12 and 2, Table 1). All of the LDN-Su1-Ph1 plants showed a multivalent at MI in some PMCs (Table 2).
To identify the wheat chromosome harboring the introgressed chromosome segment, sequential FISH and GISH were performed on LDN-Su1-Ph1. FISH with the oligo-pTa535 and oligo-pSc119.2 probes paints all wheat chromosomes and facilitates their identification (Tang et al., 2014). GISH with labeled genomic DNA of Ae. speltoides PI 369609 facilitated identification of the wheat chromosome harboring Ae. speltoides segments. Sequential FISH and GISH karyotyping of plants heterozygous for Xpsr1205-3S revealed the presence of a single recombined chromosome 3A with an Ae. speltoides segment replacing the distal portion of the long arm (Figures 2A,B). Sequential FISH and GISH karyotyping of plants homozygous for Xpsr1205-3S, showed two such chromosomes (Figures 2C,D). Thus, FISH and GISH karyotyping revealed that a single chromosome segment of Ae. speltoides was introgressed into LDN. The agreement between heterozygosity and homozygosity of the Xpsr1205-3S marker and the number of 3A chromosomes with the Ae. speltoides segments indicated that Xpsr1205-3S was located on that introgressed segment in LDN 3A.
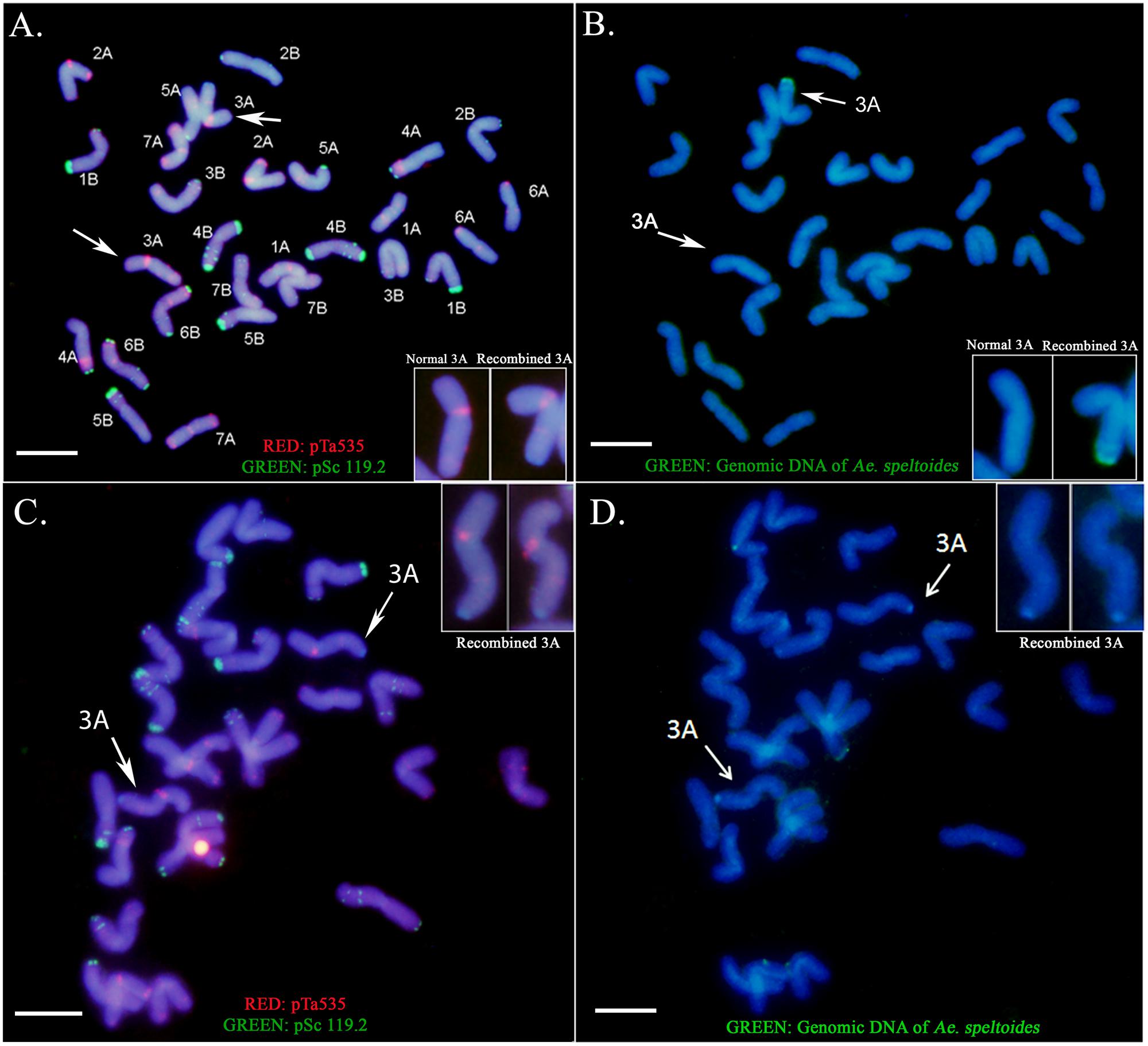
FIGURE 2. Sequential FISH and GISH karyotyping of LDN-Su1-Ph1. (A,B) The presence of a single recombined chromosome 3A with an Ae. speltoides segment replacing the distal portion of the long arm in LDN heterozygous for Xpsr1205-3S. (C,D) LDN homozygous for Xpsr1205-3S showing two recombined chromosomes 3A. The metaphase plate has only 27 chromosomes; one chromosome is outside of the field. (A,C) FISH identification of individual wheat chromosomes. (B,D) GISH identification of the recombined chromosomes. Scale bar = 10 μm.
Chromosome Pairing in Hybrids Involving CS-Su1-Ph1
Previous studies suggested that the Ae. peregrina genome does not modify Ph1 expression in wheat × Ae. peregrina hybrids (McGuire and Dvořák, 1982). If Ph1 is active in the wheat parent, there would be little chromosome pairing in wheat × Ae. peregrina hybrids but if it is inactive, extensive pairing would take place.
Seven hybrids without Xpsr1205-3S were analyzed. One hybrid without Xpsr1205-3S differed from the remaining six by having an intermediate level of chromosome pairing. The remaining six hybrids without Xpsr1205-3S had an average of 2.7 chiasmata/PMC, which was comparable to the mean chiasma number in hybrids of CS × Ae. peregrina (Table 3). In contrast, the four hybrids having Xpsr1205-3S had an average of 16.6 chiasmata per PMC (P = 0.004, t-test, N = 11 and 4). We excluded one hybrid with an exceptionally low level of chromosome pairing, although it was positive for Xpsr1205-3S (Table 3). Two hybrids from the cross ph1b × Ae. peregrina had 18.6 chiasmata/PMC, which was still higher (P = 0.012, t-test, N = 3 and 2) than the mean chiasma frequency of hybrids with Xpsr1205-3S. Overall, Ae. peregrina hybrids seemed to confirm that MAS for Xpsr1205-3S resulted in introgression of Su1-Ph1 into CS.
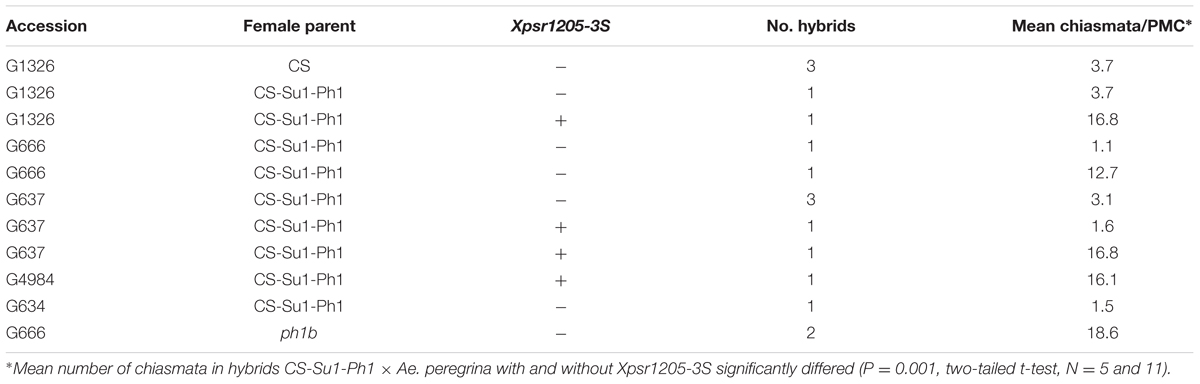
TABLE 3. Mean numbers of chiasmata per PMC in hybrids between allotetraploid Aegilops peregrina and Chinese Spring, CS ph1b, and CS-Su1-Ph1 in the presence and absence of Xpsr1205-3S.
CS-Su1-Ph1 heterozygous for Xpsr1205-3S was crossed with Ae. searsii DS5Sse(CS5A) and chromosome pairing was analyzed in progeny with and without Xpsr1205-3S. No PMC with all chromosomes paired was observed in a plant without Xpsr1205-3S (Table 4). Surprisingly, only 6.8% of PMCs showed complete pairing (a measure of pairing of the Ae. searsii 5Sse with the wheat homoeolog) in three progeny plants with Xpsr1205-3S (Table 4). This level of pairing between closely related homoeologous chromosomes was below what was expected for Ph1 expression being suppressed.
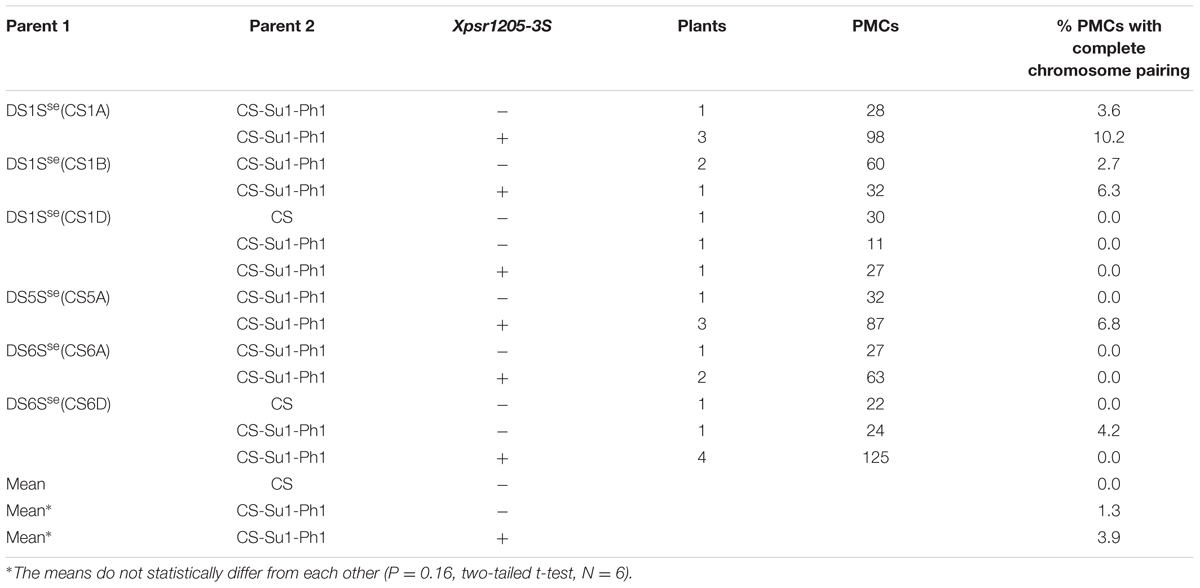
TABLE 4. Percentages of PMCs with all 42 chromosomes paired indicating pairing between wheat and Ae. searsii homoeologous chromosomes in F1 progenies from crosses of Ae. searsii DS lines (parent 1) × CS-Su1-Ph1 or CS (parent 2).
To validate this result, CS-Su1-Ph1 heterozygous for Xpsr1205-3S was crossed with DS1Sse(CS1A), DS1Sse(CS1B), DS1Sse(CS1D), DS6Sse(CS6A), and DS6Sse(CS6D). These DS lines were selected because chromosomes in these two homoeologous groups have not been reported to harbor genes affecting homoeologous chromosome pairing. DS lines for Ae. searsii chromosomes 1Sse and 6Sse were also crossed with CS as a control. PMCs that had complete chromosome pairing were rare irrespective of the presence or absence of Xpsr1205-3S (Table 4), suggesting that only minor or no suppression of Ph1 took place in the presence of Xpsr1205-3S. This finding contradicted the conclusion made on the basis of chromosome pairing in hybrids involving Ae. peregrina and indicated that Ae. peregrina genome may not be entirely neutral with respect to suppression of Ph1.
Chromosome Pairing and Recombination in the LDN Genetic Background
Because of concerns that the Ae. peregrina genome may be obscuring the true effects of the introgressed chromosome segment on the expression of Ph1, we used the CS genome in the assessment of the status of Ph1 expression in the presence of the introgresed Xpsr1205-3S. LDN-Su1-Ph1 heterozygous for Xpsr1205-3S was crossed with CS and chromosome pairing was studied in progeny. If the chromosome segment harboring Xpsr1205-3S did not also harbor Su1-Ph1, the seven D-genome monosomes would not pair with their A- and B-genome homoeologs in the pentaploid hybrids (2n = 5x = 35) and 14II + 7I would be expected at MI in most PMCs. If, on the other hand, introgression of Xpsr1205-3S did result in the introgression of Su1-Ph1 into LDN and Su1-Ph1 suppressed Ph1, the D-genome chromosomes would pair with their A and B genome homoeologoues and trivalents accompanied by fewer than 7I would be expected in PMCs of the pentaploid hybrids with Xpsr1205-3S. In the four hybrids without Xpsr1205-3S, an average of 8.0 univalents and 13.4 bivalents were observed (Table 5). In one of these hybrids, an occasional trivalent and quadrivalent was also observed; none were observed in the other three hybrids. Chromosome pairing indicated that Ph1 was active in all four hybrids. In the 11 hybrids with Xpsr1205-3S, the mean number of univalents per PMC was 5.8 (Table 5), which was significantly lower (P = 0.0002, two-tailed t = test with unequal variance) than the mean of 8.0 univalents per PMC in hybrids without Xpsr1205-3S. Concomitantly, the mean number of trivalents significantly increased to an average of 1.1 per PMC (P < 0.0001, two-tailed t = test with unequal variance). Up to four trivalents accompanied by three univalents were observed in a single PMC (Figure 3). These data were consistent with the hypothesis that Ph1 was suppressed in the pentaploid hybrids with Xpsr1205-3S and indicated that introgression of Xpsr1205-3S to LDN was accompanied by introgression of an active Su1-Ph1 allele.
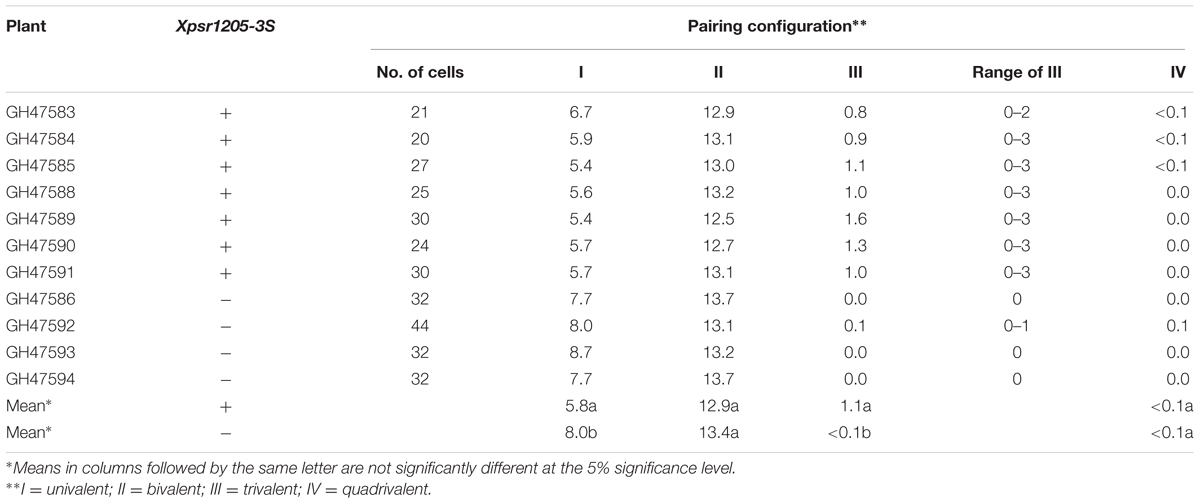
TABLE 5. Meiotic pairing in pentaploid (2n = 35) progeny from the cross LDN-Su1-Ph1 × Chinese Spring.
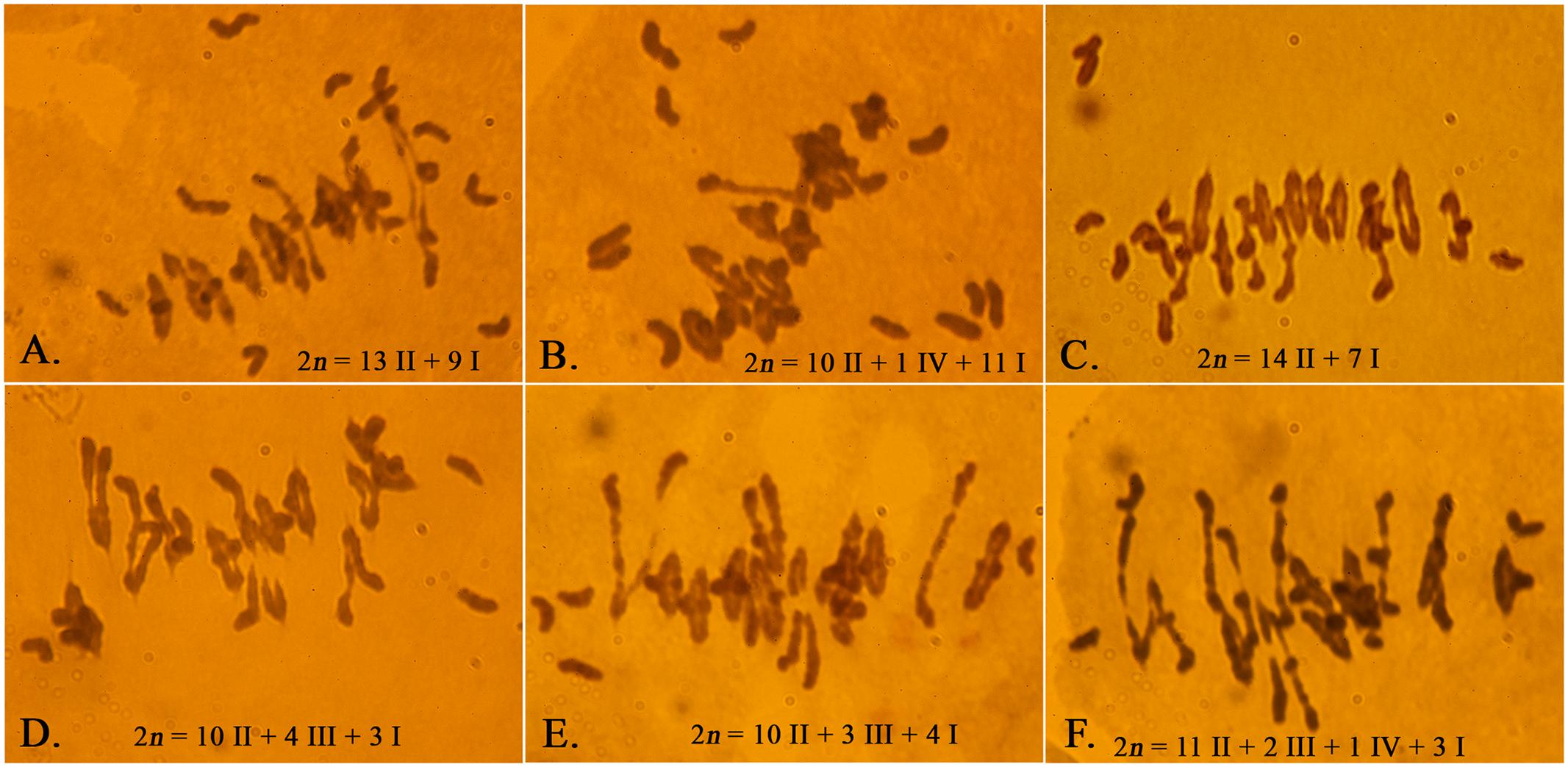
FIGURE 3. Meiotic chromosome pairing in F1 plants of LDN-Su1-Ph1 × Chinese Spring (2n = 35). (A–C) Pentaploid plants negative for Xpsr1205-3S. (D–F) Pentaploid plants positive for Xpsr1205-3S. Each PMC is from a different plant.
Additional evidence confirming suppression of Ph1 by Su1-Ph1 in the LDN genetic background was provided by meiotic pairing of Ae. searsii chromosome 5Sse with LDN chromosome 5B. A double monosomic for Ae. searsii chromosome 5Sse and LDN chromosome 5B in the LDN genetic background (2n = 28) was developed as described in Figure 4. Plants from BC1 and BC2 generation were genotyped with BE426080-5Sse and CD454867-5Sse to select plants with chromosome 5Sse and crossed with LDN-Su1-Ph1 heterozygous for Xpsr1205-3S. In F1 plants with Xpsr1205-3S, the mean numbers of PMCs with complete chromosome pairing, indicating pairing of 5Sse with 5B, was 58.3%, and was significantly higher than mean of 7.9% of PMCs in F1 lacking Xpsr1205-3S (P < 0.01, two-tailed t-test with unequal variance) (Table 6).
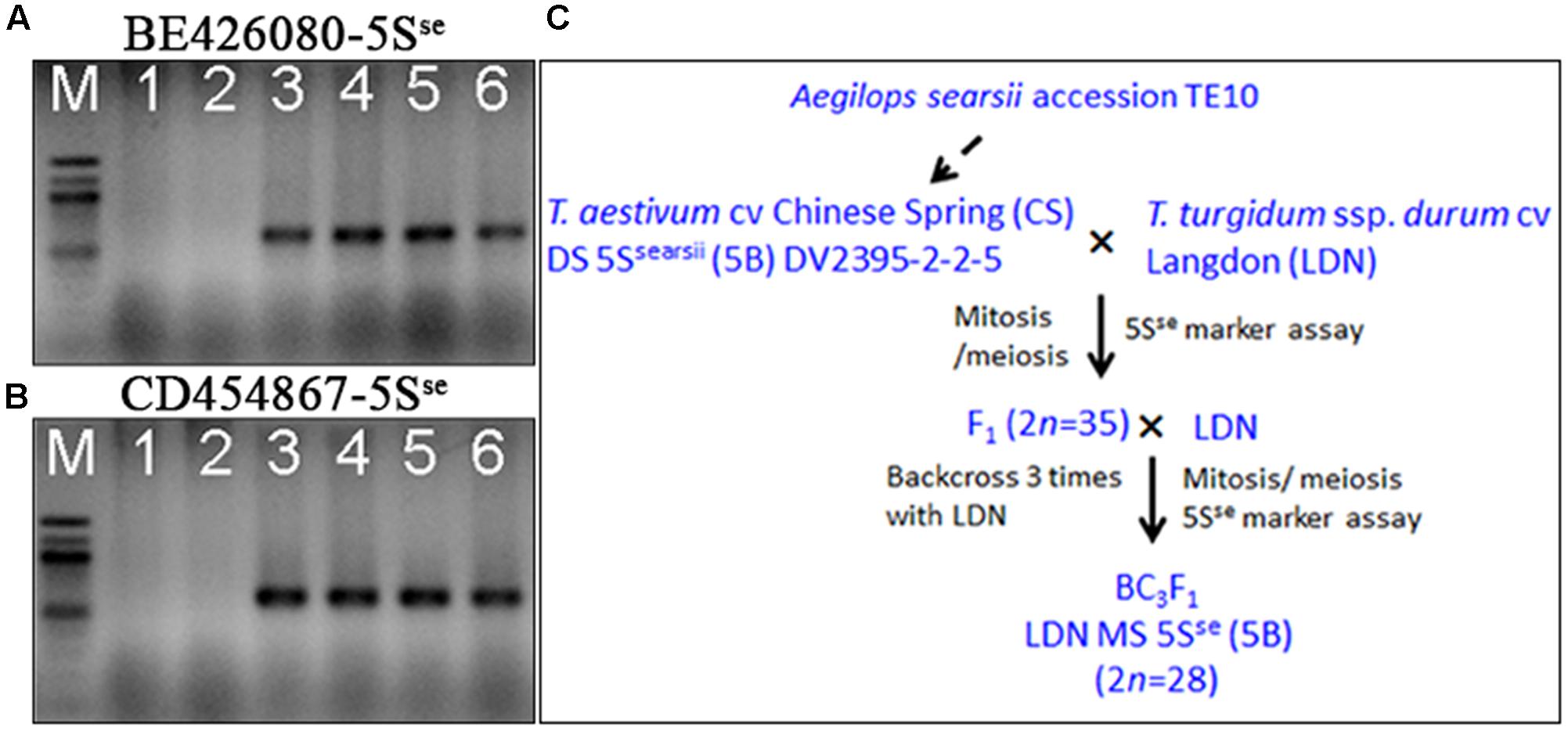
FIGURE 4. Development of a substitution line 5Sse(5B)/5B in the LDN genetic background. (A,B) Two 5Sse chromosome specific SNP markers (BE426080-5Sse and CD454867-5Sse) were developed. Lanes 1–6: LDN, CS, Ae. searsii TE10 (GH42258), Ae. searsii TE10 (GH42259), CS/Ae. searsii TE10 amphiploid (GH26353), CS/Ae. searsii TE10 amphiploid (GH26354). (C) Breeding scheme showing the development of the LDN double-monosomic substitution line 5Sse(5B)/5B. Analyses performed in each step are in black.
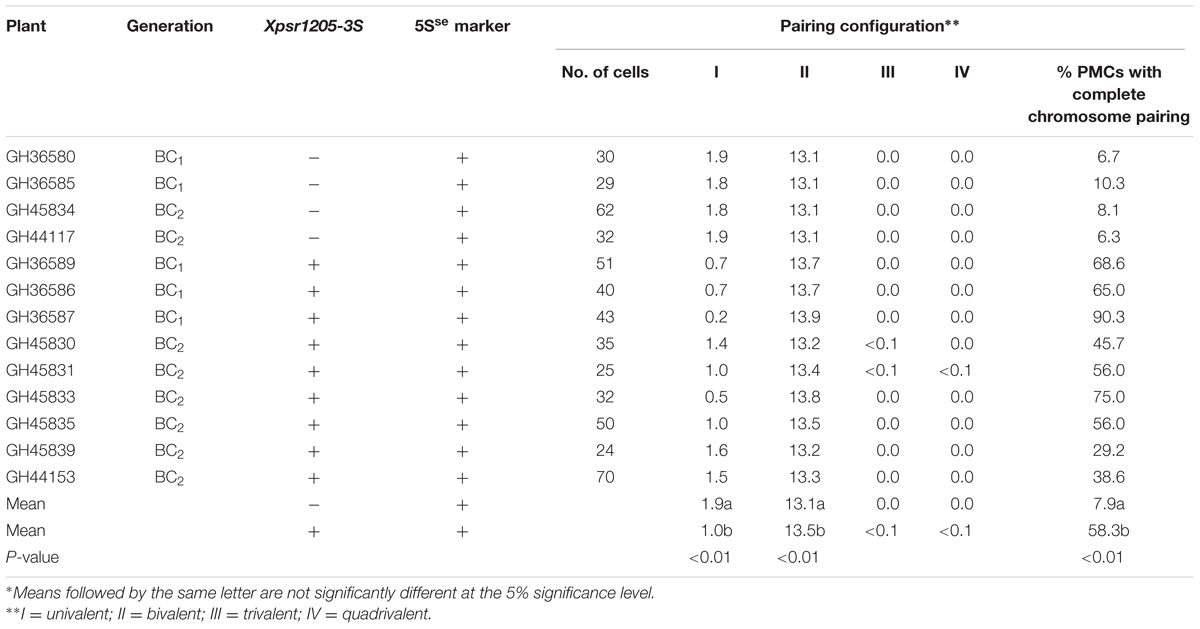
TABLE 6. Meiotic chromosome pairing in the F1 plants from the crosses of two generations (BC1 and BC2) of monosomic substitution 5Sse(LDN5B) × LDN-Su1-Ph1.
The F1 plants were selfed and also backcrossed as females to LDN-Su1-Ph1 to study recombination. Genotypes at BE426080-5Sse and CD454867-5Sse were determined in 35 F2 and 28 BC1F1 progeny. The two markers are proximally located in the Ph1 region of chromosome 5B and cosegregated at 160.6 cM on the genetic map from the cross of wild emmer (T. turgidum ssp. dicoccoides) × LDN (Jorgensen et al., 2017). Two recombined chromosomes, one in an F2 plant (GH45780) and the other in a BC1F1 plant (GH45804), were identified among 98 progeny chromosomes (Figure 5).
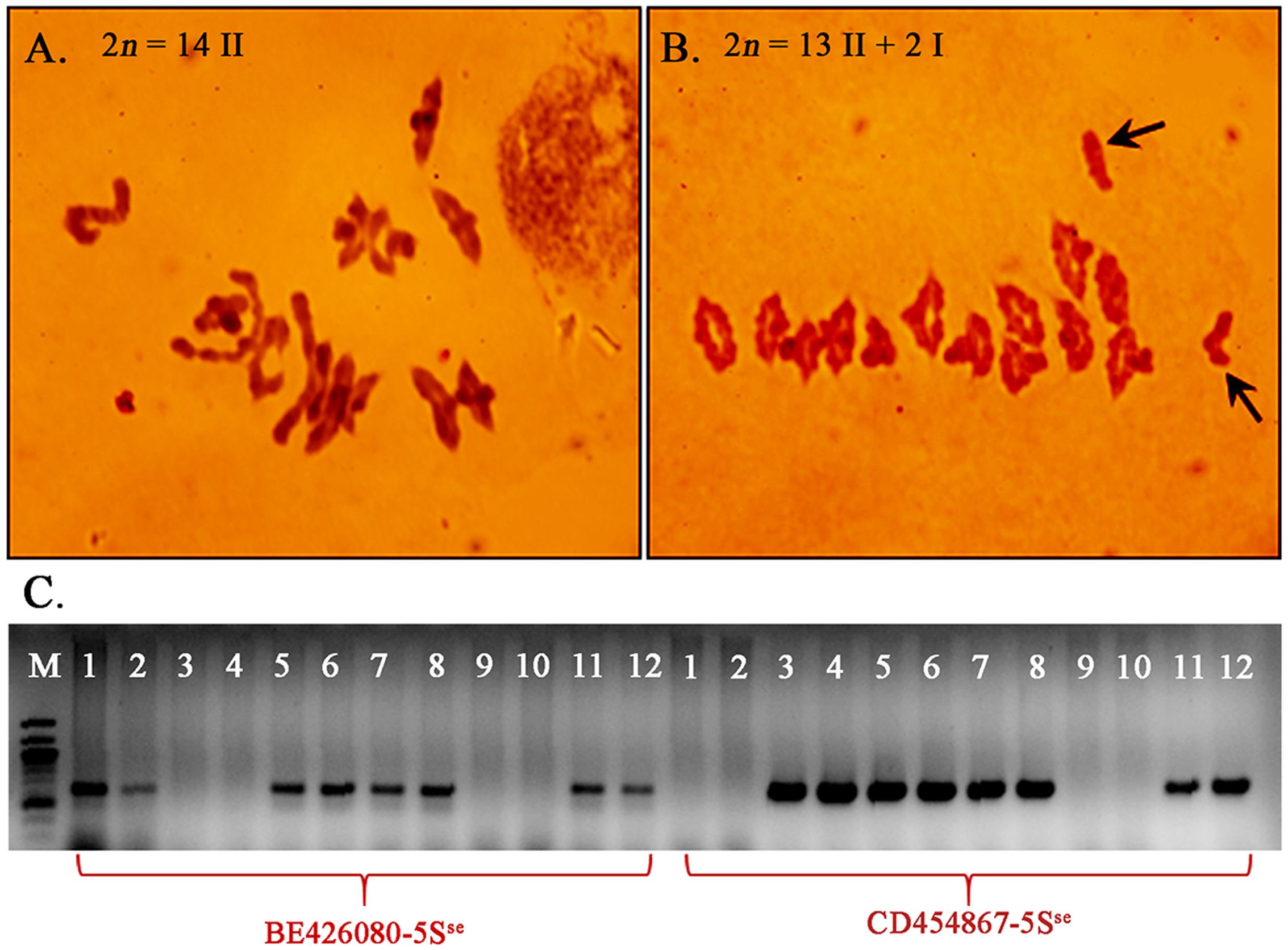
FIGURE 5. Meiotic pairing and recombination between chromosomes 5B and 5Sse induced by Su1-Ph1. Pollen mother cells showing pairing (A) and its absence (B) between chromosomes 5B and 5Sse in F1 progeny respectively with and without Xpsr1205-3S from the cross double monosomic 5Sse(5B)/5B × LDN-Su1-Ph1. Arrows in (B) indicate the unpaired univalents 5B and 5Sse. (C) Plants GH45780 and GH45804, with recombined chromosomes in the BE426080-CD454867 interval. From left, GH45780 (lanes 1–2), GH45804 (lanes 3–4), GH45837 (lanes 5–6), GH45793 (lanes 7–8), LDN (lanes 9–10), CS/Ae. searsii amphiploid (lanes 11–12).
Chromosome pairing and recombination was also assessed between distantly related homoeologous chromosomes, the L. elongatum chromosome 1E and wheat chromosome 1A, in progeny from the cross LDN-Su1-Ph1 × DS1E(LDN1A). The L. elongatum chromosome 1E did not pair with wheat chromosome 1A (N = 64) in progeny without Xpsr1205-3S but it paired with it in 6.0% PMCs (N = 83) in progeny with Xpsr1205-3S. Based on the relationship 1% MI pairing = 0.5% recombination, the length of the 1E/1A linkage group (LG) was 3% recombination. Three recombined chromosomes were identified among 63 F2 progeny (Table 7). All three crossovers were validated by genotyping of 15 to 20 F3 progeny plants. The length of the 1E/1A LG based on recombination was 2.3%, which was comparable to 3% recombination based on MI pairing (P = 0.74, 2 × 2 contingency table). Two crossovers were located within the long arm and one was located within the short arm (Table 7). Thus, also this study confirmed that Ph1 was suppressed in plants with Xpsr1205-3S in the LDN genetic background.
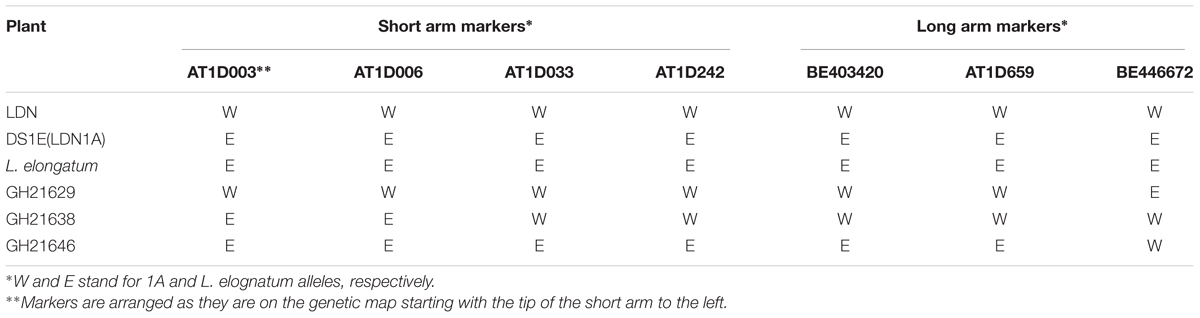
TABLE 7. Genotypes at seven SNP markers of the parental and recombined chromosomes in F2 plants of DS1E(LDN1A) × LDN-Su1-Ph1.
Discussion
Introgression of Su1-Ph1 into Wheat
Tight linkage of the Xpsr1205-3S marker to Su1-Ph1 (Dvorak et al., 2006b) was exploited here in introgression of Su1-Ph1 from Ae. speltoides into hexaploid wheat and from hexaploid wheat into tetraploid wheat. MAS was employed rather than selection for the meiotic pairing phenotype. Selection based only on meiotic phenotype would require a testcross with Ae. peregrina, or a similar tester, each backcross generation to ascertain that Ph1 is suppressed. Moreover, it will be difficult to detect the presence of a suppressor allele in the CS background if the Ph1 effect is not completely suppressed. Heterozygosity for translocations resulting from recombination between homoeologous chromosomes during backcrossing or from chromosome breakage and non-homologous end-joining due to the activity of gametocidal genes (Tsujimoto and Tsunewaki, 1984; Kota and Dvorak, 1988; Marais et al., 2010) could potentially lead to mistaking such multivalent pairing for homoeologous pairing.
The CS × Ae. speltoides hybrid treated with colchicine had dehiscent anthers and produced two octoploid seeds, indicating that it was male fertile but the two octoploid progeny plants were male sterile. They were therefore backcrossed as females with DS3E(CS3B) for two generations. Since L. elognatum chromosome 3E partially suppresses Ph1 (Dvořák, 1987), it is therefore theoretically possible that the suppressor of Ph1 introgressed in CS-Su1-Ph1 and LDN-Su1-Ph1 was the 3E suppressor rather than Su1-Ph1. This possibility is very unlikely for the following reasons. First, chromosome 3E has a weak suppressor, which is located in the short arm (Dvořák, 1987), whereas Su1-Ph1 is a strong suppressor, which is located in the long arm. Second, the Xpsr1205-3S primers for PCR amplification were Ae. speltoides specific, and when DS3E(CS3B) was used as a template, no amplification was obtained. Therefore, using Xpsr1205-3S in introgression could not have introgressed a 3E segment in place of the targeted Ae. speltoides chromosome segment. Third, in situ hybridization showed that in plants with Xpsr1205-3S, the terminal segment of 3A was replaced by an Ae. speltoides chromosome segment. Finally, the LDN-Su1-Ph1 introgression lines were subjected to nine generations of backcrossing, first to CS and then to LDN. The probability that a recombined chromosome bearing the 3E suppressor was present in the LDN-Su1-Ph1 lines was <0.002.
Five approaches were employed to test the hypothesis that Su1-Ph1 was introgressed along with introgression of Xpsr1205-3S with the following results. (1) D-genome chromosomes frequently paired in trivalents with their A- and B-genome homoeologs in the pentaploid LDN-Su1-Ph1 × CS hybrids that acquired Xpsr1205-3S but not in those that did not acquire Xpsr1205-3S. (2) Chromosome 5Sse paired with LDN chromosome 5B in 58.3% of PMCs if Xpsr1205-3S was present but only in a few percent of PMCs if Xpsr1205-3S was absent. Recombined 5Sse/5B chromosomes were recovered in progeny of plants with Xpsr1205-3S. (3) L. elongatum chromosome 1E paired in the LDN genetic background with chromosome 1A if Xpsr1205-3S was present but did not pair with it if Xpsr1205-3S was absent. Recombined 1E/1A chromosomes were recovered in progeny. (4) A mean of 16.6 chiasmata per PMC was observed in F1 hybrids CS-Su1-Ph1 × Ae. peregrina if Xpsr1205-3S was present but only 2.7 chiasmata/PMC were observed if Xpsr1205-3S was absent. The last approach (5) provided evidence contradictory to that provided by approaches (1) to (4). Single Ae. searsii chromosomes 1Sse, 5Sse, and 6Sse paired with wheat homoeologs in only a few percent of PMCs in progenies involving CS-Su1-Ph1 in the presence of Xpsr1205-3S. Thus, the presence of the introgressed Xpsr1205-3S consistently resulted in suppression of Ph1 in the LDN genetic background but the results were inconsistent in the CS genetic background.
Sequential FISH and GISH showed that LDN-Su1-Ph1 plants with Xpsr1205-3S harbored an Ae. speltoides chromosome segment replacing a distal portion of the long arm of chromosome 3A. The correct orientation of the segment in the arm suggests that the translocation originated by recombination between LDN chromosome 3A and Ae. speltoides chromosome 3S. We name the chromosome as T(3AL;3SL)Dv1.
The LDN T(3AL;3SL)Dv1 chromosome was introgressed into LDN from CS-Su1-Ph1. If the T(3AL;3SL)Dv1 chromosome harbored Su1-Ph1 in LDN, the chromosome must have also harbored the Su1-Ph1 in CS-Su1-Ph1. To account for these puzzling contradictions, we hypothesize that Su1-Ph1 normally interacts with at least one other gene in Ae. speltoides that was lost during introgression. We further hypothesize that this complementary gene is absent in the CS genome but present in the LDN and Ae. peregrina genomes.
Evidence for the presence of the complementary gene in LDN was provided by extensive homoeologous chromosome pairing in the LDN-Su1-Ph1 × CS pentaploid hybrids with Xpsr1205-3S. The fact that no homoeologous chromosome pairing took place in the same hybrids devoid of Xpsr1205-3S suggested that the LDN gene affects chromosome pairing only when complemented by Su1-Ph1.
Aegilops peregrina (genomes UUSvSv) and tetraploid wheat (genomes AABB) each have one genome pair closely related to the S genome of Ae. speltoides (Dvorak, 1998; Badaeva et al., 2004) and each could have possessed this complementary gene we postulate to exist in Ae. speltoides. Ae. peregrina accession G637 may be polymorphic for the gene, as the hybrid with Xpsr1205-3S but with low pairing may suggest.
Chromosome pairing in the LDN-Su1-Ph1 and CS-Su1-Ph1 introgression lines is consistent with the hypothesis that the complementary gene(s) is present in LDN but not in CS. Occasional multivalents from heterogenetic chromosome pairing were observed in all LDN-Su1-Ph1 plants. In contrast, no multivalents were observed in the CS-Su1-Ph1 plants.
Since tetraploid and hexaploid wheat share the A and B genomes, and since there has been extensive gene flow between tetraploid and hexaploid wheat (Dvorak et al., 2006a; Akhunov et al., 2010), it is very likely that hexaploid wheat is polymorphic for the complementary gene. A search for the gene in T. aestivum will require the development of a complementation assay that would indicate the presence of homoeologous chromosome pairing in hybrids involving Xpsr1205-3S.
Molecular Nature of Su1-Ph1 and Its Relationship to ZIP4-1 and Zip4-B2
The Su1-Ph1 locus and ZIP4-1 gene, the likely source of the Ph1 gene (Rey et al., 2017), are both located on the long arm of chromosome 3A. Does the spatial and functional relationship of Su1-Ph1 and ZIP4-1 indicate equivalence of the two loci? The locations of these loci on the reference genome sequences of wild emmer (Avni et al., 2017) and Ae. tauschii (Luo et al., 2017) suggest that they are not equivalent. Su1-Ph1 was mapped between Xpsr1205-3S and EST locus XBF497740 on two independent maps. Su1-Ph1 and XBF497740 were 0.2 and 13 cM proximal to Xpsr1205-3S, respectively (Dvorak et al., 2006b). Orthologous Xpsr1205 loci are on the 3A, 3B, and 3D pseudomolecules at 712,850,941, 786,992,681, and 591,216,484 bp, respectively (Figure 6). The orthologous XBF497740 loci are about 30 Mb proximal to Xpsr1205 in each pseudomolecule (Figure 6) but the ZIP4-A1 and ZIP4-B1 and ZIP4-D1 loci, which are at collinear locations on pseudomolecules 3A, 3B and 3D, are about 30 Mb further proximal to the XBF497740 marker and about 70 Mb proximal to Xpsr1205. It is therefore very unlikely that ZIP4-1 and Su1-Ph1 are equivalent. Whether ZIP4-B1 is actually the complementary locus detected in LDN or whether Su1-Ph1 regulates the expression of ZIP4-B1 and ZIP4-B2 must be investigated.
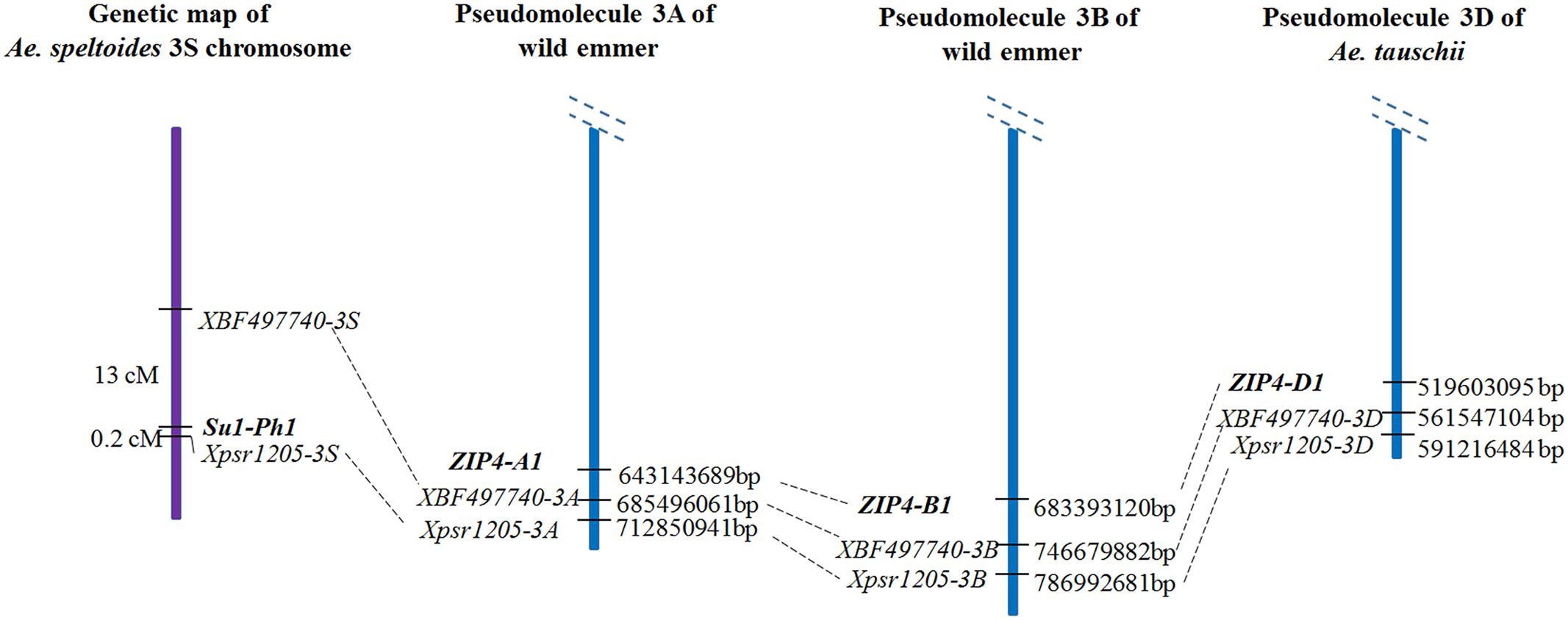
FIGURE 6. Molecular nature of Su1-Ph1 and relationship to ZIP4-1. Su1-Ph1 was flanked by Xpsr1205-3S (0.2 cM) and EST locus XBF497740 (13 cM) (Dvorak et al., 2006b). The order of the three loci on the 3A and 3B wild emmer pseudomolecules (Avni et al., 2017) and the 3D Ae. tauschii pseudomolecule (Luo et al., 2017) is as shown. ZIP4-1 is about 30 Mb proximal to the XBF497740 marker and about 70 Mb proximal to Xpsr1205, which indicates that it is very unlikely that ZIP4-1 and Su1-Ph1 are equivalent.
Comparison of chromosome pairing in hybrids of frameshift mutants of ZIP4-B2 (Cad1691 and Cad0348) × Ae. peregrina (Rey et al., 2017) with the hybrids of CS-Su1-Ph1 × Ae. peregrina suggests that Ph1 expression was not entirely abolished by the Cad1691 and Cad0348 mutations. The hybrids had an average of 12.2 chiasmata/PMC (Rey et al., 2017) whereas those involving CS-Su1-Ph1 had an average of 16.6 chiasmata/PMC (P < 0.001, t-test, N = 3 and 2). Both sets of hybrids had lower chiasma frequency than the hybrids involving the ph1b deletion mutation (18.6 chiasmata/PMC). The average number of chiasmata in hybrids involving the ZIP4-B2 mutants was similar to the exceptional hybrid involving CS-Su1-Ph1 × Ae. peregrina accession G666, which showed an intermediate level of chromosome pairing (12.7 chiasmata/PMC). The mutants of ZIP4-B2 did not show multivalents at MI in meiosis (Rey et al., 2017), which is consistent with partial activity of Ph1. Evidence that ZIP4-B2 is equivalent to Ph1 hinges on chromosome pairing in the hybrids with Ae. peregrina. As we have learned, the Ae. peregrina genome can obscure the actual effect of genes affecting Ph1, and it would be prudent to provide additional evidence that Ph1expression was abolished in Cad1691 and Cad0348 ZIP4-B2 mutants.
Practical Utility of the Introgressed Su1-Ph1
Since the presence of heterozygous Xpsr1205-3S in the LDN-Su1-Ph1 × CS F1 hybrids elicited homoeologous chromosome pairing, Su1-Ph1 must be dominant. The dominant epistasis of Su1-Ph1 over Ph1 makes it a very flexible means of inducing recombination between homoeologs. To achieve recombination between an alien chromosome and a wheat homoeolog, the alien chromosome should be substituted for a wheat homoeolog and the substitution line should be crossed with an introgression line harboring the T(3AL;3SL)Dv1 chromosome. The introgression line supplies both the Ph1 suppressor and the wheat homoeologs for recombination with the alien chromosome. This strategy was illustrated here by targeting recombination between homoeologous chromosomes 5Sse with 5B and 1E with 1A.
There are however two caveats. One is that Su1-Ph1 will suppress Ph1 only if one of the parents contributes the complementary gene to F1 progeny. This requirement is satisfied by the LDN-Su1-Ph1 introgression lines but not by the CS-Su1-Ph1 introgression lines. The alien chromosome substitution lines should therefore be in the LDN genetic background, as was done here to achieve recombination of 5Sse with 5B and 1E with 1A. It is also possible to use hexaploid alien chromosome substitutions and rely on the induction of recombination between the homoeologs in the pentaploid hybrids, but that strategy has not been tested.
The other caveat is that Su1-Ph1 does not suppress Ph1 completely. The chiasma frequency in CS-Su1-Ph1 × Ae. peregrina F1 hybrids was significantly lower than the average chiasma frequency in the ph1b × Ae. peregrina F1 hybrids. Therefore, Su1-Ph1 will be the most effective in the manipulation of closely related homoeologous chromosomes, although, as illustrated here, even more distantly related chromosomes, such as L. elongatum and wheat, could be recombined.
Author Contributions
JD, HL, and AD conceived and designed the experiments. HL, KD, and JD performed the experimental work. JD, HL, and KD analyzed the data. JD, HL, KD, M-CL, WJ, and AD discussed the findings and interpreted the results. JD and HL wrote the first draft of the paper. All authors have read and approved the final draft.
Funding
This work was supported in part by the National Science Foundation of United States (grant no. IOS1212591), USDA Plant Genome (grant no. 2006-01161), NIFA Hatch Program no. 1002302, and BARD Project no. IS-4829-15.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors thank Patrick McGuire (University of California, Davis, Davis, CA, United States) for critical reading and valuable suggestions for the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02163/full#supplementary-material
Footnotes
References
Akhunov, E. D., Akhunova, A. R., Anderson, O. D., Anderson, J. A., Blake, N., Clegg, M. T., et al. (2010). Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics 11:702. doi: 10.1186/1471-2164-11-702
Avni, R., Nave, M., Barad, O., Baruch, K., Twardziok, S. O., Gundlach, H., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97. doi: 10.1126/science.aan0032
Badaeva, E., Amosova, A., Samatadze, T., Zoshchuk, S., Shostak, N., Chikida, N., et al. (2004). Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Syst. Evol. 246, 45–76. doi: 10.1007/s00606-003-0072-4
Chen, P. D., Tsujimoto, H., and Gill, B. S. (1994). Transfer of Ph (I) genes promoting homoeologous pairing from Triticum speltoides to common wheat. Theor. Appl. Genet. 88, 97–101. doi: 10.1007/BF00222400
Dover, G. A., and Riley, R. (1972). Prevention of pairing of homoeologous meiotic chromosomes of wheat by an activity of supernumerary chromosomes of Aegilops. Nature 240, 159–161. doi: 10.1038/240159a0
Dvořák, J. (1972). Genetic variability in Aegilops speltoides affecting homoeologous pairing in wheat. Can. J. Genet. Cytol. 14, 371–380. doi: 10.1139/g72-046
Dvořák, J. (1987). Chromosomal distribution of genes in diploid Elytrigia elongata that promote or suppress pairing of wheat homoeologous chromosomes. Genome 29, 34–40. doi: 10.1139/g87-006
Dvorak, J. (1998). “Genome analysis in the Triticum-Aegilops alliance,” in Proceedings of the 9th International Wheat Genetics Symposium, Saskatoon, SK, 8–11.
Dvorak, J., Akhunov, E. D., Akhunov, A. R., Deal, K. R., and Luo, M.-C. (2006a). Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol. Biol. Evol. 23, 1386–1396.
Dvorak, J., Deal, K. R., and Luo, M. C. (2006b). Discovery and mapping of wheat Ph1 suppressors. Genetics 174, 17–27.
Dvorak, J., Terlizzi, P., Zhang, H. B., and Resta, P. (1993). The evolution of polyploid wheats: identification of the A genome donor species. Genome 36, 21–31. doi: 10.1139/g93-004
Dvorak, J., and Zhang, H. B. (1990). Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes. Proc. Natl. Acad. Sci. U.S.A. 87, 9640–9644. doi: 10.1073/pnas.87.24.9640
Friebe, B., Tuleen, N., and Gill, B. (1995). Standard karyotype of Triticum searsii and its relationship with other S-genome species and common wheat. Theor. Appl. Genet. 91, 248–254. doi: 10.1007/BF00220885
Giorgi, B. (1978). A homoeologous pairing mutant isolated in Triticum durum cv. Cappelli. Mutat. Breed. Newsl. 11, 4–5.
Griffiths, S., Sharp, R., Foote, T. N., Bertin, I., Wanous, M., Reader, S., et al. (2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439, 749–752. doi: 10.1038/nature04434
Jauhar, P., Almouslem, A., Peterson, T., and Joppa, L. (1999). Inter- and intragenomic chromosome pairing in haploids of durum wheat. J. Hered. 90, 437–445. doi: 10.1093/jhered/90.4.437
Jauhar, P., Riera-Lizarazu, O., Dewey, W., Gill, B., Crane, C., and Bennett, J. (1991). Chromosome pairing relationships among the A, B, and D genomes of bread wheat. Theor. Appl. Genet. 82, 441–449. doi: 10.1007/BF00588597
Jauhar, P. P., and Peterson, T. S. (2011). Cytological and molecular characterization of homoeologous group-1 chromosomes in hybrid derivatives of a durum disomic alien addition line. Plant Genome 4, 102–109. doi: 10.3835/plantgenome2011.01.0002
Jenczewski, E., and Alix, K. (2004). From diploids to allopolyploids: the emergence of efficient pairing control genes in plants. Crit. Rev. Plant Sci. 23, 21–45. doi: 10.1080/07352680490273239
Jorgensen, C., Luo, M.-C., Ramasamy, R., Dawson, M., Gill, B., Korol, A., et al. (2017). A high-density genetic map of wild emmer wheat from the Karaca dag region provides new evidence on the structure and evolution of wheat chromosomes. Front. Plant. Sci. 8:1798. doi: 10.3389/fpls.2017.01798
Kihara, H. (1944). Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Agric. Hortic. 19, 13–14. doi: 10.1016/j.jgg.2011.07.002
Kimber, G., and Athwal, R. (1972). A reassessment of the course of evolution of wheat. Proc. Natl. Acad. Sci. U.S.A. 69, 912–915. doi: 10.1073/pnas.69.4.912
Kimber, G., and Riley, R. (1963). The relationships of the diploid progenitors of hexaploid wheat. Can. J. Genet. Cytol. 5, 83–88. doi: 10.1139/g63-012
Koo, D.-H., Liu, W., Friebe, B., and Gill, B. S. (2016). Homoeologous recombination in the presence of Ph1 gene in wheat. Chromosoma 126, 531–540. doi: 10.1007/s00412-016-0622-5
Kota, R. S., and Dvorak, J. (1988). Genomic instability in wheat induced by chromosome 6B (S) of Triticum speltoides. Genetics 120, 1085–1094.
Li, H., Gill, B. S., Wang, X., and Chen, P. (2011). A Tal-PhI wheat genetic stock facilitates efficient alien introgression. Genet. Resour. Crop Evol. 58, 667–678. doi: 10.1007/s10722-010-9609-x
Li, H., Wang, C., Fu, S., Guo, X., Yang, B., Chen, C., et al. (2014). Development and discrimination of 12 double ditelosomics in tetraploid wheat cultivar DR147. Genome 57, 89–95. doi: 10.1139/gen-2013-0153
Luo, M.-C., Gu, Y. Q., Puiu, D., Wang, H., Twardziok, S. O., Deal, K. R., et al. (2017). Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502. doi: 10.1038/nature24486
Luo, M.-C., Gu, Y. Q., You, F. M., Deal, K. R., Ma, Y. Q., Hu, Y., et al. (2013). A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc. Natl. Acad. Sci. U.S.A. 110, 7940–7945. doi: 10.1073/pnas.1219082110
Marais, G., Bekker, T., Eksteen, A., Mccallum, B., Fetch, T., and Marais, A. (2010). Attempts to remove gametocidal genes co-transferred to common wheat with rust resistance from Aegilops speltoides. Euphytica 171, 71–85. doi: 10.1007/s10681-009-9996-2
McFadden, E. S., and Sears, E. R. (1946). The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37, 107–116. doi: 10.1093/oxfordjournals.jhered.a105594
McGuire, P. E., and Dvořák, J. (1982). Genetic regulation of heterogenetic chromosome pairing in polyploid species of the genus Triticum sensu lato. Can. J. Genet. Cytol. 24, 57–82. doi: 10.1139/g82-007
Rey, M.-D., Martín, A. C., Higgins, J., Swarbreck, D., Uauy, C., Shaw, P., et al. (2017). Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol. Breed. 37:95. doi: 10.1007/s11032-017-0700-2
Riley, R. (1960). The diploidisation of polyploid wheat. Heredity 15, 407–429. doi: 10.1038/hdy.1960.106
Riley, R., and Chapman, V. (1958). Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182, 713–715. doi: 10.1038/182713a0
Riley, R., Chapman, V., and Miller, T. (1973). “The determination of meiotic chromosome pairing,” in Proceedings of the 4th International Wheat Genetics Symposium, eds E. R. Sears and L. M. S. Sears (Colombia, MO: University of Missouri), 731–738.
Sears, E., and Okamoto, M. (1958). Intergenomic chromosome relationships in hexaploid wheat. Proc. 10th Int. Congr. Genet. 2, 258–259. doi: 10.1093/aob/mcm331
Sears, E. R. (1977). An induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 19, 585–593. doi: 10.1139/g77-063
Tang, Z., Yang, Z., and Fu, S. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Tsujimoto, H., and Tsunewaki, K. (1984). Gametocidal genes in wheat and its relatives. I. Genetic analyses in common wheat of a gametocidal gene derived from Aegilops speltoides. Can. J. Genet. Cytol. 26, 78–84. doi: 10.1139/g84-013
Tuleen, N., and Hart, G. (1988). Isolation and characterization of wheat-Elytrigia elongata chromosome 3E and 5E addition and substitution lines. Genome 30, 519–524. doi: 10.1139/g88-087
Keywords: homoeologous chromosome pairing, in situ hybridization, MAS, Ph1, ZIP4
Citation: Li H, Deal KR, Luo M-C, Ji W, Distelfeld A and Dvorak J (2017) Introgression of the Aegilops speltoides Su1-Ph1 Suppressor into Wheat. Front. Plant Sci. 8:2163. doi: 10.3389/fpls.2017.02163
Received: 28 October 2017; Accepted: 07 December 2017;
Published: 20 December 2017.
Edited by:
Roberto Papa, Università Politecnica delle Marche, ItalyReviewed by:
Karolina Susek, Institute of Plant Genetics (PAN), PolandTomás Naranjo, Complutense University of Madrid, Spain
Copyright © 2017 Li, Deal, Luo, Ji, Distelfeld and Dvorak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jan Dvorak, jdvorak@ucdavis.edu
 Hao Li
Hao Li Karin R. Deal1
Karin R. Deal1 Ming-Cheng Luo
Ming-Cheng Luo Assaf Distelfeld
Assaf Distelfeld Jan Dvorak
Jan Dvorak