Comparative chloroplast genomics of three species of Bulbophyllum section Cirrhopetalum (Orchidaceae), with an emphasis on the description of a new species from Eastern Himalaya
- Published
- Accepted
- Received
- Academic Editor
- Victoria Sosa
- Subject Areas
- Biodiversity, Conservation Biology, Genomics, Plant Science, Taxonomy
- Keywords
- Bulbophyllum, Chloroplast genome, New species, Orchidaceae, Phylogenetic position
- Copyright
- © 2023 Li et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2023. Comparative chloroplast genomics of three species of Bulbophyllum section Cirrhopetalum (Orchidaceae), with an emphasis on the description of a new species from Eastern Himalaya. PeerJ 11:e14721 https://doi.org/10.7717/peerj.14721
Abstract
Background
Chloroplast (cp) genomes are useful and informative molecular markers used for species determination and phylogenetic analysis. Bulbophyllum is one of the most taxonomically complex taxa in Orchidaceae. However, the genome characteristics of Bulbophyllum are poorly understood.
Methods
Based on comparative morphological and genomic analysis, a new species Bulbophyllum pilopetalum from eastern Himalaya belonging to section Cirrhopetalum is described and illustrated. This study used chloroplast genomic sequences and ribosomal DNA (nrDNA) analysis to distinguish the new Bulbophyllum species and determine its phylogenetic position. An additional phylogenetic analysis was conducted using 74 coding sequences from 15 complete chloroplast genomes from the genus Bulbophyllum, as well as nrDNA sequences and two chloroplast DNA sequences from 33 Bulbophyllun species.
Results
The new species is morphologically similar to B. pingnanense, B. albociliatum, and B. brevipedunculatum in vegetative and floral morphology, but it can be distinguished by its ovate-triangle dorsal sepal without a marginal ciliate. The chloroplast genome of the new Bulbophyllum species is 151,148 bp in length, and includes a pair of inverted repeats (IRs) of 25,833 bp, a large single-copy region (LSC) of 86,138 bp, and a small single-copy region (SSC) of 13,300 bp. The chloroplast genome includes 108 unique genes encoding 75 proteins, 30 tRNAs, and four rRNAs. Compared with the cp genomes of its two most closely-related species, B. pingnanense and B. albociliatum, this chloroplast genome exhibited great interspecific divergence and contained several Indels that were specific to the new species. The plastid tree showed that B. pilopetalum is most closely-related to B. pingnanense. The phylogenetic tree based on combined nrDNA and chloroplast DNA sequences indicated that section Cirrhopetalum was monophyletic and B. pilopetalum was a member of this section.
Discussion
The taxonomic status of the new species is strongly supported by cp genome data. Our study highlights the importance of using the complete cp genome to identify species, elucidate the taxonomy, and reconstruct the phylogeny of plant groups with complicated taxonomic problems.
Introduction
Bulbophyllum Thouars is one of the largest orchid genera with 2,000–2,200 species that are distributed all over the pantropical regions, especially in the Paleotropics (Pridgeon et al., 2014). Bulbophyllum species have rounded pseudobulbs bearing one or two leaves and small flowers that often have colored sepals that are larger than the petals. This genus represents one of the most diverse genera in the Orchidaceae family, exhibiting a wide variety of growth forms and floral characteristics (Gamisch & Comes, 2019). The genus of Bulbophyllum has a complicated taxonomic history. Bulbophyllum was initially established in 1822 (Thouars, 1822). Nearly 50 independent and closely related genera were described, e.g., Cirrhopetalum Lindl., Drymoda Lindl., Monomeria Lindl., Trias Lindl., and Sunipia Lindl. (Pridgeon et al., 2014; Seidenfaden, 1979). All of these allied genera have been combined with and separated from Bulbophyllum over the years. Molecular phylogenetic research recently supported a broad definition of Bulbophyllum that included all related genera (Chase et al., 2015; Gravendeel, Fischer & Vermeulen, 2014).
The Cirrhopetalum alliance in the genus Bulbophyllum is derived from the genus Cirrhopetalum, which is characterized by its sub-umbellate raceme inflorescence and significantly elongated lateral sepals that are twisted inward near base and connate (Garay, Hamer & Siegerist, 1994; Seidenfaden, 1973). It is a species-rich group, with ca. 150-240 species, mainly distributed in Asia (Seidenfaden, 1973; Vermeulen, 2014). Vermeulen (2014) considered it to be monophyletic and divided into 10 sections: Acrochaene (Lindl.) J. J. Verm., Biflorae Gary, Hamer & Siegerist, Blepharistes J. J. Verm., Brachyantha Rchb.f., Cirrhopetaloides Garay, Hamer & Siegerist, Cirrhopetalum (Lindl.) Rchb. f., Emarginatae Garay, Hamer & Siegerist, Eublepharon J. J. Verm., Recurvae Garay, Hamer & Siegerist, and Rhytionanthos (Garay, Hamer & Siegerist) J. J. Verm. However, Hu et al. (2020) combined DNA and floral characters to reconstruct the phylogenetic relationship in the Asian Cirrhopetalum alliance and further indicated that the majority of traditional sectional classifications were shown to be highly artificial. In the latest revision of Bulbophyllum in Flora of China, sections Umbellata Bentham & J. D. Hooker, Corymbosa (Blume) Aver., Cirrhopetalum, “Zhonghuazu”, and “Suihuazu”, which include 46 species, are involved in the Cirrhopetalum alliance (Chen & Vermeulen, 2009).
New species and records in the Cirrhopetalum alliance from China have been discovered more recently, e.g., B. picturatum (Loddiges) H. G. Reichenbach, B. pingnanense J. F. Liu, S. R. Lan & Y. C. Liang (Liu et al., 2016), B. yunxiaoense M. H. Li, J. F. Liu & S. P. Chen (Li et al., 2017), B. yongtaiense J. F. Liu, S. R. Lan & Y. C. Liang (Liu et al., 2018), B. reflexipetalum J. D. Ya, Y. J. Guo & C. Liu (Ya et al., 2019), B. wendlandianum (Kraenzl.) Dammer (Jin, Li & Ye, 2019). However, taxonomists still face difficulties in identifying the Cirrhopetalum alliance at the species level, and taxonomic uncertainties at the sectional level are majorly problematic and require revision.
During botanical explorations in the eastern Himalayas, we discovered a distinct species of Bulbophyllum from southeastern Xizang. Its sub-umbellate inflorescence and lateral sepals that are longer than its dorsal sepals strongly suggested that it should be assigned to the Cirrhopetalum alliance. Detailed examination of its morphology by living individuals and a survey of the literature (Hsu & Chung, 2008; Chen & Vermeulen, 2009; Liu et al., 2016) showed that it was morphologically related to B. pingnanense, B. albociliatum (Tang S. Liu & H. Y. Su) Seidenf., and B. brevipedunculatum T. C. Hsu & S. W. Chung, but it is distinguished from them by its smooth dorsal sepals.
Many new species have been described in recent years, and molecular markers have greatly enhanced our understanding of species delimitation (Li et al., 2017; Liu et al., 2018). Chloroplast (cp) genome sequences have contributed to solving phylogenetic and taxonomic problems in many plant groups and provided barcodes for identifying species (Niu et al., 2018; Pfanzelt, Albach & von Hagen, 2019). It is possible to obtain a complete cp genome for plants using next-generation sequencing techniques (Wilkinson et al., 2017). However, there is a lack of genetic and molecular data for this diverse genus of Bulbophyllum. The complete cp genome may be used to examine sequence divergence that could help resolve taxonomic uncertainties in the Bulbophyllum genus.
In this study, we sequenced and performed a comparative analysis of the complete cp genome of the new species and its closely related species, B. pingnanense and B. albociliatum. Furthermore, molecular phylogenetic analysis using the complete cp genome and nrDNA internal transcribed spacer (ITS) was conducted to determine the phylogenetic position of this new species. This study aims to confirm that our newly collected specimen is an undescribed species based on cp genome sequence divergence and use whole cp genome data to resolve taxonomic uncertainties within the Cirrhopetalum alliance.
Materials & Methods
Morphological analyses
Morphological characteristics of the new species were studied based on the study of living materials. Images of flowering plants were taken in the field. In addition, the morphological study included a comparison of the new species with other species, based on the Flora of China (Chen & Vermeulen, 2009) and a bibliography of related species (Hsu & Chung, 2008; Liu et al., 2016). A total of 13 diagnostic characteristics of the new species were compared to those of three closely related species. Voucher Specimens were deposited in the Herbarium of Xishuangbanna Tropical Botanical Garden (HITBC), Chinese Academy of Sciences, while the living individuals were preserved in the nursery.
New botanical taxonomic name
The electronic version of this article in Portable Document Format (PDF) will represent a published work according to the International Code of Nomenclature for plants (ICN), and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone. In addition, new names contained in this work which have been issued with identifiers by IPNI will eventually be made available to the Global Names Index. The IPNI LSIDs can be resolved and the associated information viewed through any standard web browser by appending the LSID contained in this publication to the prefix “http://ipni.org/”. The online version of this work is archived and available from the following digital repositories: PeerJ, PubMed Central SCIE, and CLOCKSS.
Sampling, DNA extraction and sequencing
Fresh leaves of B. pilopetalum (Voucher: Luo et al. 3521) and B. albociliatum (Voucher: Xu & Wang 202106) were collected from the living collections at the nursery of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences and Yunnan Fengchunfang Biotechnology Co. Ltd. Additionally, we downloaded the available complete cp genomes of 12 Bulbophyllum species and DNA sequences (cpDNA: mat K and psb A-trn H; nrDNA: ITS) of 30 species of the Cirrhopetalum alliance of Bulbophyllum from GenBank (Table S1) to analyze genomic characteristics and phylogenetic relationships. The methods of both DNA extraction and sequencing were followed Tang et al. (2021).
Chloroplast genome assembly and annotation
GetOrganelle1.7.5 was used for de novo cp genome and the nrDNA sequence (18S-ITS1−5.8S-ITS2-26S) assembly with default parameters (Jin et al., 2020). CPGAVAS2 (Shi et al., 2019) and GeSeq (Tillich et al., 2017) were used to annotate the assembled cp genome using default parameters to predict protein-coding, rRNA, and tRNA genes. The sequence identity and annotation results were checked and the manual corrections and codon positions were adjusted by comparing them with Bulbophyllum species present in the database using GeneiousPrime 2021. The length of the whole plastome, number of genes, categories of genes, and GC content were analyzed in GeneiousPrime. The genome map of the species was illustrated with the help of OGDRAW (Greiner, Lehwark & Bock, 2019). The annotated cp genome and nrDNA sequences were submitted to GenBank (Table S1).
Comparative genomic and phylogenetic analysis
Comparative genomic analysis, including cp genomes sequence divergence, mutations and indels, the borders, and measurement of the nucleotide diversity value (Pi), was conducted as previously described in Tang et al. (2021). The nucleotide identity of the cp genomes of all three Bulbophyllum species was calculated using GenieousPrime.
We selected the complete cp genomes of the two newly sequenced Bulbophyllum species, 13 Bulbophyllum species, and two species from Dendrobium (Table S1) downloaded from GenBank to reconstruct the phylogenetic tree. Phylogenetic analysis was performed by 74 protein-coding sequences of the cp genome, using Maximum Parsimony (MP), Maximum Likelihood (ML), Bayesian (BI) analyses. The methods of constructing phylogenetic tree were previously described in Tang et al. (2021). Additionally, to estimate the systematic position of the new species within the Cirrhopetalun alliance, we used nrDNA ITS and two cp DNA sequences (matK, psb A-trnH) from 30 species of the Cirrhopetalum alliance available from GenBank to reconstruct their phylogenetic relationships (Table S1). Dendrobium species were used as the outgroups. The phylogenetic relationship analysis was performed using ML methods combined with nr DNA and cp DNA sequences.
Results
Taxonomy
Etymology. The specific epithet “pilopetalum” refers to the petal margins dense with white ciliate.
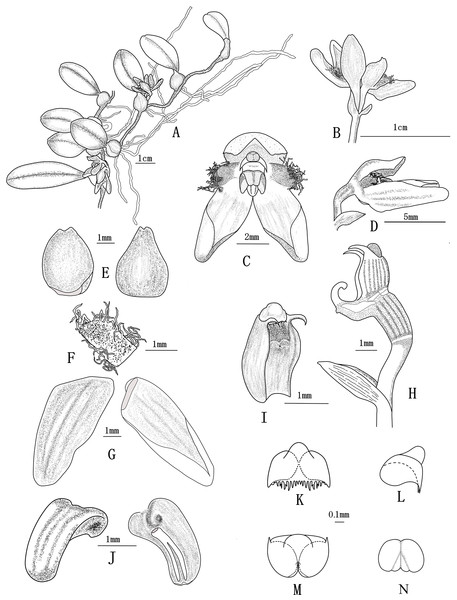
Figure 1: Bulbophyllum pilopetalum M. K. Li, J. P. Deng & Y. Luo. (All drawn from Y. Luo et al. 3521 by J. P. Deng).
(A) Flowering plant; (B) inflorescence; (C) flower, frontal view; (D) flower, side view; (E) dorsal sepal, adaxial and abaxial top view; (F) petal, adaxial top view; (G) lateral sepal, adaxial and abaxial top view; (H) Column and ovary, side view; (I) column, bottom view; (J) lip, side view and bottom view; (K–M) anther cap, frontal view, top view and bottom view; (N) pollinia, frontal view.Figure 2: Bulbophyllum pilopetalum. M. K. Li, J. P. Deng & Y. Luo. (Photos from Y. Luo et al. 3521 by J. P. Deng in Bomi, Tibet, China).
(A) Habit; (B–C) plant and inflorescence; (D) sepals, petals and lip frontal view; (E) flower, frontal view; (F) dorsal sepal, abaxial top view; (G) lateral sepal, abaxial top view; (H) lip, side view; (I) petal, top view; (J) flower and floral bract, side view; (K) column and ovary, side view; (L) lip and Column foot, bottom view; (M) pollinia, frontal view; (N-O) anther cap, top view and bottom view.Holotype. CHINA. XIZANG: Bomi County, Tongmai Village, on the trunks of trees or sometimes on rocks in the humid evergreen broad-leaved forest, 2100 m, Mar. 10, 2021, Y. Luo, M. K. Li & L. Tang 3521, fl. [holotype (HITBC0074007) & isotype (HITBC0074008), HITBC).
Diagnosis. Similar to B. brevipedunculatum, B. albociliatum, and B. pinganense, but differs by having a shorter peduncle, triangle dorsal sepal with margin entire, oblong petals, and divergent lateral sepals twisted slightly near the apex.
Description. Short-creeping epiphyte. Rhizome ca. 0.7 mm in diam. Pseudobulbs 1.2–2 cm apart on rhizome, subglobose or ovoid, 5–7 mm, 3–5 mm in diam., young enveloped by brownish scarious sheaths, old with fibrous remnants at the base, with a terminal leaf. Leaf sessile; blade oblong to ovate-lanceolate, 1– 4 × 0.7–0.8 cm, rigid, base cuneate, apex obtuse. Scape from the base of pseudobulb, 3–5 mm, slightly shorter than pseudobulb, sub-umbel 1- or 2-flowered; peduncle less than 5 mm, enclosed in 2 or 3 membranous sheaths; floral bracts ovate-lanceolate, ca. 3 mm, apex shortly acute. Pedicel and ovary ca. 5 mm. Flowers orangeish, with orange sepals and red petals. Sepals free, thickly textured; dorsal sepal concave, ovate-triangular, 3.5–4 mm × ca. 2 mm, margin entire, apex acute and beaked; lateral sepals oblong, 7–8 mm × 2–3 mm, twisted inward near the base, free and slightly divergent, margin entire, apex obtuse. Petals oblong, 1.5–2 mm × ca. one mm, margin densely long white ciliate, apex rounded; lip orange, recurved, oblong-lanceolate, ca. 2 × 1.1 mm, basal half grooved, base attached to the end of column foot by a moveable joint, apex obtuse; disk nearly smooth adaxially. Column pale white, ca. 1.5 mm, stout, with a distinct foot, conspicuously winged; stelidia filiform, acute, ca. 0.5 mm; foot curved, ca. 1.5 mm, with free part ca. 0.8 mm; anther cap subglobose, orange, fimbriate at apex; pollinia 2, ovoid, each with one deep groove dividing pollinium into two unequal particles, without appendage. Ovary 5–6 ribbed, glabrous, ca. 2 mm long, 0.8 mm in diam.
Conservation status. Bulbophyllum pilopetalum is known only from the type locality, and only two small populations of ca. 10 individual plants within a narrow area were discovered. The appropriate data on the abundance and distribution of this species is lacking. It can be estimated IUCN Red List status—Data Deficient (DD) (IUCN 2012).
Notes. Morphologically, B. pilopetalum is allied to some taxa from the eastern coast of China, namely B. pingnanense, B. albociliatum, and B. brevipedunculatum by its dwarf plants, pseudobulbs separated by rhizomes about 1 to 2 cm, orangeish flowers and petals with ciliate margin, elongated lateral sepals twisted inward near the base. These three species are placed under sect. Cirrhopetalum (Chen & Vermeulen, 2009; Liu et al., 2016). Our plant has similar characteristics and should also belong to this section. However, it differs from other species by having an ovate-triangular dorsal sepal with a conspicuous beak and entire margin, and oblong petals. The differences between these four related species are shown in Table 1. B. pilopetalum may be most closely related to B. brevipedunculatum, which has a short scape and 1–3 flowers, but our plant can be distinguished by its oblong lateral sepals, open divergent. B. pilopetalum differs from B. pingnanense and B. albociliatum by having a shorter scape, lateral sepals, and fewer flowers.
| Characters | B. pilopetalum | B. brevipedunculatuma | B. albociliatumb | B. pingnanensec |
|---|---|---|---|---|
| Pseudobulbs apart | 1.2–2 cm | 1–2 cm | 2 cm | 0.6–2.5 cm |
| Pseudobulb | subglobose or ovoid, 5–7 mm, 3–5 mm in diam. | elongate ovoid, 7–10 mm, 3–5 mm in diam. | elongate-ovoid, 10–13 mm, 5–7 mm in diam. | obvate-elliptic, 0.5–1.7 cm, 3–6 mm in diam. |
| Leaf | oblong to ovate-lanceolate, 1–4 × 0.7–0.8 cm, apex obtuse, emarginate | oblong to linear-oblong, 1–7 cm × 0.7–0.8 cm, apex obtuse to retuse | oblong, oblanceolate or obovate, 2–4 cm × 0.7–1 cm, apex obtuse or rounded | oblong to linear-oblong, 1.8–6.6 × 0.6–1.2 cm, apex obtuse to retuse |
| Scape | scape slightly shorter than pseudobulb, 3–5 mm, 1–2 flower | scape shorter than pseudobulb, 5–7 mm, 1- 3 flowered | scape much longer than pseudobulb, ca. 4–6 cm, 5–6 flower | scape much longer than pseudobulb, ca. 1.1 cm, 3–5 flower |
| Floral bract | ovate-lanceolate, ca. 3 mm | elongate triangular, ca. 3 mm | narrowly triangular, ca. 3 mm | triangular, ca.2–3 mm |
| Pedicel and ovary | 5 mm | 4–5 mm | 5 mm | 4 mm |
| Flower | orangeish, with red petals | reddish | rubescent, with reddish yellow lateral sepals | orange red |
| Dorsal sepal | concave, ovate-triangular, 3.5–4 × ca. 2 mm, entire, apex acute and beaked | strongly concave, elliptic, ca. 3.5 mm × 2 mm, apex acuminate, short white ciliate | rubescent, strongly concave, elliptic, 3–4 mm × 2 mm, apex rounded, long white ciliate | concave, ovate, abaxially papillose, 5 × 3 mm, apex obtuse or acute margins long white ciliate, |
| Lateral sepal | oblong, 7–8 × 2–3 mm, apex obtuse, open divergent | near rectangular, 5–7 mm × 2–3 mm, apex mucronate, often connate | lanceolate, 7–9 mm × 2 mm, incurved along margins, often connate | laceolate, 10–12 × 2 mm, apex acute, open divergent or connate |
| Petal | oblong, ca. 1.5–2 mm × 1 mm, margins densely with white ciliate, apex rounded | elliptic, ca. 2 × 1.2 mm, white ciliate, apex rounded, | elliptic, ca. 2 mm × 1.2 mm, apex rounded, long white ciliate | ovate, ca. 2.7–3.0 × 1.2–2.0 mm, margins long white ciliate, apex round |
| Lip | orange, oblong-lanceolate, ca, 2 × 1.1 mm, apex rounded obtuse, | dark red, horn-like, ca. 2 mm, apex rounded obtuse | rubescent, horn-like, ca. 1.5 mm, recurved, adaxial surface of lip partly papillose | ovate-triangular, ca. 3 mm, abaxially deeply grooved |
| Column | pale white, 1.5 mm | pale yellow, ca. 1.5 mm, | yellow, ca. 1.5 mm | yellow, subterete, ca. 1–2 mm |
| Anther cap | subglobose | cordate | cordate | subglobose |
Characteristics of Bulbophyllum chloroplast genome
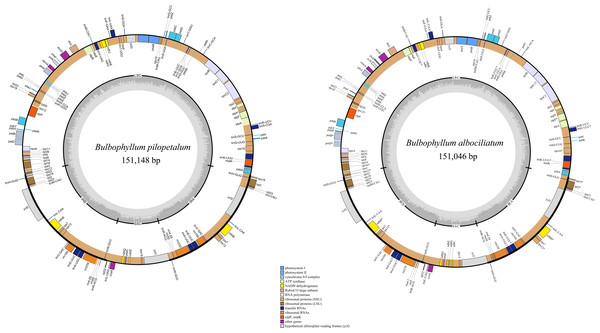
The lengths of the cp genomes of B. pilopetalum and its two closely related species, B. albociliatum and B. pingnanense (MW822749), are 151,148 bp, 151,224 bp, and 151,046 bp, respectively (Fig. 3, Table 2). Three cp genomes displayed a typical quadripartite structure and contained a pair of inverted repeats (IRs, 25,833 to 25,855 bp), separated by a large single copy region (LSC, 85,956 to 86,138 bp) and a small single copy region (SSC, 13,300 to 13,441 bp), respectively (Table 2). The overall GC contents of B. pilopetalum was 36.9% and that of both B. pingnanense and B. albociliatum was 37%. Three of the cp genomes each consistently contained 109 unique genes which were arranged in the same order across the genomes, including 75 protein-coding (CDS), 30 tRNA, and four rRNA genes (Table 3). The gene map was shown in Fig. 3. Fourteen genes (eight CDS and six tRNA) contained one intron in three Bulbophyllum species and three genes (ycf3, clpP, and rps12) contained two introns (Table 3).
Figure 3: Chloroplast genome map for Bulbophyllumpilopetalum M. K. Li, J. P. Deng & Y. Luo (A) and B. albociliatum (T. S. Liu et H. J. Su) Seidenf. (B).
Genes located outside the outer rim are transcribed in a counterclockwise direction, whereas genes inside the outer rim are transcribed in a clockwise direction. The colored bars indicate known different functional groups. The dashed gray area in the inner circle shows the percentage GC contents of the corresponding genes. LSC, SSC, and IR denote large single copy, small single copy, and inverted repeat, respectively.| B. pilopetalum | B. pingnanense | B. albociliatum | |
|---|---|---|---|
| genome size (bp) | 151,148 | 151,224 | 151,046 |
| LSC length (bp) | 86,138 | 86,017 | 85,956 |
| SSC length (bp) | 13,300 | 13,441 | 13,384 |
| IR length (bp) | 25,833 | 25,855 | 25,853 |
| Number of genes | 109 | 109 | 109 |
| Protein-coding genes | 75 | 75 | 75 |
| tRNA genes | 30 | 30 | 30 |
| rRNA genes | 4 | 4 | 4 |
| GC content (%) | 36.9 | 37 | 37 |
| GC content in LSC (%) | 34.4 | 34.4 | 34.5 |
| GC content in SSC (%) | 29.1 | 29.2 | 29.2 |
| GC content in IR (%) | 43.2 | 43.2 | 43.2 |
| GC content in Protein-coding (%) | 37.9 | 37.9 | 37.8 |
| GC content in tRNA (%) | 52.8 | 53 | 53 |
| GC content in rRNA IR (%) | 54.9 | 54.9 | 54.9 |
| Group of genes | Gene names |
|---|---|
| 1 Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| 2 Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| 3 Cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN |
| 4 ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI |
| 5 NADH dehydrogenase | ndhA*, ndhB*(×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| 6 RubisCO large subunit | rbcL |
| 7 RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 |
| 8 Ribosomal proteins(SSU) | rps2, rps3, rps4, rps7(×2), rps8, rps11, rps12**(×2), rps14, rps15, rps16*, rps18, rps19(×2) |
| 9 Ribosomal proteins(LSU) | rpl2(×2), rpl14, rpl16*, rpl20, rpl22, rpl23(×2), rpl32, rpl33, rpl36 |
| 10 Other genes | clpP**, matK, accD, ccsA, infA, cemA |
| 11 Proteins of unknown function | ycf1, ycf2(×2), ycf3**, ycf4 |
| 12 Ribosomal RNAs | rrn4.5S(×2), rrn5S(×2), rrn16S(×2), rrn23S(×2) |
| 13 Transfer RNAs | trnA-UGC*(×2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC*, trnH-GUG(×2), trnI-CAU(×2), trnI-GAU*(×2), trnK-UUU*, trnL-CAA(×2), trnL-UAA*, trnL-UAG, trnM-CAU(×2), trnN-GUU(×2), trnP-UGG, trnQ-UUG, trnR-ACG(×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(×2), trnV-UAC*, trnW-CCA, trnY-GUA |
Comparative genomic analysis
Sequence divergence
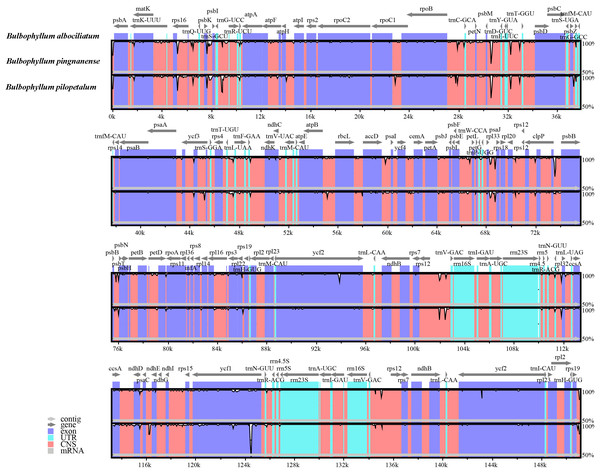
The comparative sequence analysis of three Bulbophyllum species revealed that the cp sequences were highly conserved across the three taxa with a few variable regions (Fig. 4). The sequences were more conserved in IR regions, whereas, most of the divergence detected were found in LSC and SSC regions (Fig. 4).
Figure 4: Alignment of the cp genome sequences of three Bulbophyllum species, with B. albociliatum as a reference.
The X-axis corresponds to coordinates within the cp genome. The Y-axis shows the percentage identity in the range 50% to 100%.Mutations and indels
The number of mutations and indel events was compared between the cp genomes of B. pilopetalum and its two closely related species, respectively. There were 1,952 mutations and 445 indels calculated between B. pilopetalum and B. albociliatum, with the highest indel rate of 0.33 indels per 100 bp in the intergenic regions, while the highest mutation rate was 1.70 mutations per 100 bp in introns (Table 4). There were 1,575 mutations and 239 indels calculated between B. pilopetalum and B. pingnanense, with the highest indel rate of 0.22 indels per 100 bp in introns, while the highest mutation rate was 1.25 mutations per 100 bp in the intergenic regions (Table 4).
| B. pilopetalum vs B. albociliatum | B. pilopetalum vs B. pingnanense | |||||
|---|---|---|---|---|---|---|
| CDS | IGS | intron | CDS | IGS | intron | |
| Indels | 125 | 262 | 58 | 91 | 95 | 53 |
| Mutations | 438 | 1142 | 372 | 477 | 993 | 105 |
| Total length | 72,632 | 79,255 | 21,939 | 72,209 | 79,235 | 23,828 |
| indels/100 bp | 0.17 | 0.33 | 0.26 | 0.13 | 0.12 | 0.22 |
| mutations/100 bp | 0.60 | 1.44 | 1.70 | 0.66 | 1.25 | 0.44 |
Notes:
- CDS
-
protein-coding genes
- IGS
-
intergenic spacers
We further analyzed the details of indels (indel size>10 bp) of the three Bulbophyllum species based on multiple whole cp genome alignments. DNA indels were found in 36 positions in at least one of the cp genomes, predominately in the intergenic regions, as well as in introns or in the coding regions. B. pilopetalum and B. pingnanense shared 11 indels, and B. pilopetalum and B. albociliatum shared 11 indels as well. There were 14 unique indels in B. pilopetalum, including ycf1 (111 bp, coding region), psbI-atpA (50 bp), ycf3 (29 bp, intron), psbZ-rps14 (24 bp), rpl12-rpl32 (20 bp), ycf1-rpl2 (20 bp), and psbM-petD (12 bp) (Table S2).
Border contraction and extension
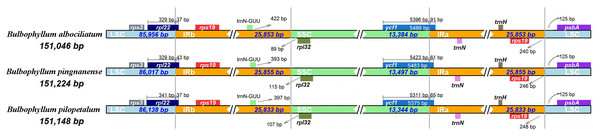
We analyzed the IR/single copy (SC) region border positions and their adjacent genes in the three Bulbophyllum cp genomes (Fig. 5). In all cp genomes of the three Bulbophyllum species, the genes rpl22, rps19, trnN-GUU, rpl32, ycf1, and psbA were located at the junction of the LSC/IRb, IRb/SSC, SSC/IRa, and IRa/LSC borders (Fig. 5). The border regions of cp genomes were similar across the three Bulbophyllum species. There was 37 to 43 bp extension of rpl22 gene into the IRb region. There was 393–422 bp between trnN- GUU and the LSC/IRa border, while the rpl32 generated a distance of 89–115 bp to another LSC/IRa junction. The ycf1 gene crossed the SSC/IRa junction expanding 61–91 bp to IRa regions in three species. There was 240–248 bp between rps19 and the LSC/IRa border, and the psbA generated a distance of 125 bp to another LSC/IRa junction.
Figure 5: Comparison of the borders between neighboring genes and junctions of LSC, SSC, and IR regions of the chloroplast genomes in three Bulbophyllum species.
Boxes above or below the main line indicate genes adjacent to borders.Nucleotide diversity and identity
The nucleotide diversity value (Pi) of the cp genome was calculated using DnaSP between the new species and its closely related species to understand their sequence divergence. The Pi values between B. pilopetalum and B. albociliatum ranged from 0 to 0.0179, which were higher compared to those between B. pilopetalum and B. pingnanense, with the Pi values ranging from 0 to 0.0150 (Fig. S1). The nucleotide identity of the cp genome was 99.124% between B. pilopetalum and B. albociliatum, and 99.229% between B. pilopetalum and B. pingnanense.
Phylogenetic position of new species
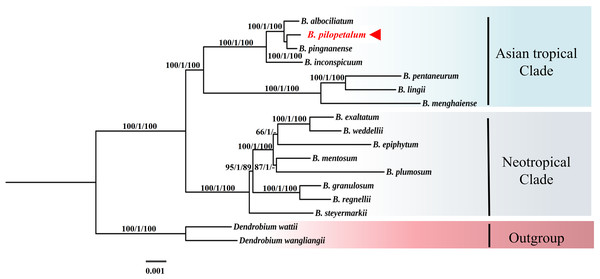
In the phylogenetic tree of 74 protein-coding genes of the cp genome using ML, MP, and BI analyses (Fig. 6), the Bulbophyllum genus was monophyletic and formed two clades, the Asian tropical and Neotropical Bulbophyllum. The Asian tropical clade formed two subclades: three species from B. sect. Macrocaulia (Blume) Aver., and B. inconspicuum Maxim. (Native to Japan) was a sister to these three closely related species, which strongly supported that these three species were monophyletic. Within this clade, the sister relationship between B. pilopetalum and B. pingnanense was highly supported.
Figure 6: Phylogenetic position of Bulbophyllum pilopetalum based on maximum likelihood (ML), Bayesian inference (BI) and maximum parsimonious (MP) analysis of 74 protein-coding chloroplast genes from Bulbophyllum species.
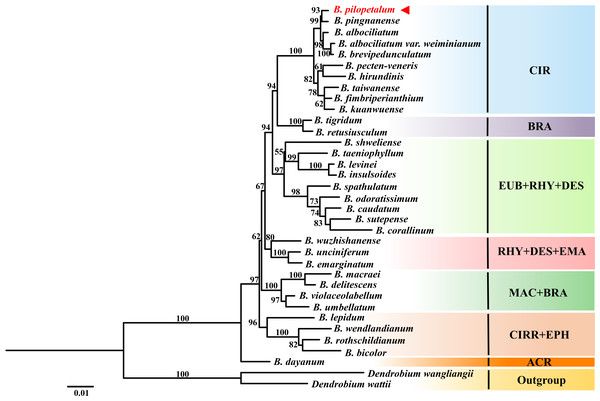
The numbers above branches represent bootstrap percentage (BP) of ML/posterior probability (PP) of BI /BP of MP (“-” indicates a BP < 90%).ML analyses were conducted based on nrDNA ITS and two cp DNA sequences from 33 species of the Cirrhopetalum alliance (Fig. 7). Phylogenetic analysis showed seven clades were grouped, but they were not well consistent with sectional classification by Chen & Vermeulen (2009) and Vermeulen (2014). The clade included B. dayanum from sect. Acrochaene formed the base of the Cirrhopetalum alliance. The ML trees showed a strongly supported clade (CIR) grouping 10 species from sect. Cirrhopetalum together. Within this clade, B. pilopetalum and B. pingnanense were placed together as sisters. Six species from sect. Brachyantha were divided into two subclades. Four other clades were recognized with strong support, but each included species from two or three sections (Fig. 7).
Figure 7: Phylogenetic position of Bulbophyllum pilopetalum based on Maximum Likelihood analysis of the combined nuclear (ITS) and chloroplast DNA sequence (matK and psbA-trnH).
The numbers above branches are ML bootstrap support values (BP); sectional classification of taxa (according to Chen & Vermeulen, 2009; Vermeulen, 2014) is indicated by three-letter abbreviations following the taxon name: ACR: sect. Acrochaene, BRA: sect. Brachyantha, CIR: sect. Cirrhopetalum, CIRR: sect. Cirrhopetaloides, DES: sect. Desmosanthes, EPH: sect. Ephippium, EUB: sect. Eublepharon, EMA: sect. Emarginatae, MAC: sect. Macrostylida, RHY: sect. Rhytionanthos.Discussion
Evidence for species delimitation based on chloroplast genomes
The cp genome sequence provided comprehensive and adequate information to better understand the phylogeny and improve species identification efficiency (Niu et al., 2018; Pfanzelt, Albach & von Hagen, 2019). Comparative cp genomic analysis of three species from B. sect. Macrocaulia showed cp genomes of sect. Macrocaulia species had similar structure and gene content and shared a number of indels, which mainly contributed to its monophyly (Tang et al., 2021). Cp genomes of eight Neotropical Bulbophyllum species provide good resolution for the sectional classification of Neotropical Bulbophyllum (Zavala-Páez et al., 2020). The comparison of the cp genome structure between B. pilopetalum, B. albociliatum, and B. pingnanense revealed that the gene composition, gene structure, and the number of genes were found to be similar, and the differences between the border contraction and extension were not significant. The nucleotide identity of the cp genome between the three species ranged from 99.124–99.270%, with a slightly higher similarity of 99.229% between B. pilopetalum and B. pingnanense. The cp genome showed high sequence divergence between B. pilopetalum and its two closely related species: Pi value of 0−0.0179 between B. pilopetalum and B. albociliatum and 0−0.0150 between B. pilopetalum and B. pingnanense. The mutations and indels were frequently found between the cp genomes of three species, whereas 14 unique indels (indel size>10 bp) were found in B. pilopetalum. The differences in cp genome structure distinguish B. pilopetalum from other closely related species, further supporting its status as a distinct species. The indels specific to this species can be used as diagnostic DNA sequence characteristics (Table S2).
The new species could be placed in B. sect. Cirrhopetalum
Section Cirrhopetalum belongs to the core Cirrhopetalum alliance (Hu et al., 2020). However, its taxonomic status and circumscription remain controversial. Sect. Cirrhopetalum is characterized by sub-umbellate raceme, lateral sepals longer than the dorsal sepal, twisting and connected lateral sepals, and hairy dorsal sepal and petals (Seidenfaden, 1979). Several researchers have proposed different classifications of the sect. Cirrhopetalum. For example, sect. Cirrhopetalum was listed with 42 species in Flora of China (Tsi, Chen & Luo, 1999), but was later revised to 17 species (Chen & Vermeulen, 2009). The molecular phylogeny of Hu et al. (2020) showed that the previous sect. Cirrhopetalum was polyphyletic and divided into two major clades, B and D7 (Hu et al., 2020). To determine the phylogenetic position of the new species, we selected nrDNA ITS and cp DNA sequences from eight Bulbophyllum species from a highly supported subclade of sect. Cirrhopetalum (Subclade D7), regarded as reliable members of sect. Cirrhopetalum according to Hu et al. (2020), and 23 other representative species from the Cirrhopetalum alliance, to reconstruct the phylogenetic tree. The phylogenetic analysis strongly supported that the new species is closely related to B. pingnanense within the sect. Cirrhopetalum clade (Fig. 7). The cp genome also supported that the new species is a sister to B. pingnanense (Fig. 6).
However, the new species is most closely and morphologically similar to B. brevipedunculatum. This is perhaps because morphological characteristics have not been fully understood between these species. In the phylogenetic tree based on nrDNA ITS and cp DNA sequences, sect. Cirrhopetalum was supported as a monophyletic clade, excluding B. lepidum, B. wendlandianum, and B. rothschildianum (Fig. 7). Within this clade, B. albociliatum, B. brevipedunculatum, and B. albociliatum var. weiminianum T.P. Lin & L. L. K. Huang formed a subclade, indicating that the three taxa were closely related. B. brevipedunculatum was treated as a variety of B. albociliatum: B. albociliatum var. brevipedunculatum (T. C. Hsu & S.W. Chung) W. M. Lin (Lin & Wang, 2014). The phylogenetic analysis in this study seems to support Lin’s taxonomic treatment. However, other clades do not correspond with the taxa elucidated in Chen & Vermeulen’s previous classification, suggesting that the current sectional classifications of the Cirrhopetalum alliance were only artificial and are in need of taxonomic revision.
The CIR clade in our study (Fig. 7) or the CIRRII (subclade D7) in the study of Hu et al. (2020) is well supported because a monophyletic group could be regarded as the circumscription of sect. Cirrhopetalum. Furthermore, although the sampling in the phylogenetic analysis of cp genome in this study was limited, the species B. inconspicuum distributed in Japan also clustered into a clade with our species. All these species have similar vegetative features, yellow, orange, or reddish flowers and dorsal sepals/petals with margins ciliate. These morphological features, such as dwarf epiphyte, short sub-umbellate inflorescence, yellowish flower, and ciliate sepals and petals, might be the diagnostic characteristics for sect. Cirrhopetalum. Dorsal sepal/petal with ciliate margin was considered to be the ancestral characteristics in the Cirrhopetalum alliance. However, our species has dorsal sepals with entire margin, which is a gradual transition from smooth to ciliate (or ciliate to smooth) dorsal sepal that might occur in this species. Other members included in previous sect. Cirrhopetalum, determined by Chen & Vermeulen (2009) and Vermeulen (2014) (e.g., species in the B clade of Bulbophyllum phylogeny of Hu et al. (2020), which was previously placed in sect. Cirrhopetalum), should be considered to revise their taxonomic status. In order to further revise sect. Cirrhopetalum, the Cirrhopetalum alliance, even the Bulbophyllum genus, we need to integrate comprehensive morphological characteristics (e.g., vegetative leaf and pseudobulbs, pollinia, and seeds) and more suitable molecular markers (e.g., cp genomes).
Supplemental Information
Sampling and accession information for Bulbophyllum and Dendrobium species in this study
The characters of indels (< 10 bp) in the chloroplast genomes of three Bulbophyllum species. “1” insersion; “0” deletion
Comparative analysis of nucleotide diversity (Pi) values between the cp genome sequences of Bulbophyllum species
(A–B) Pi values of coding genes and intergenic regions in the LSC, SSC, and IR regions between B. pilopetalum and B. albociliatum (C–D) Pi values of coding genes and intergenic regions in the LSC, SSC, and IR regions between B. pilopetalum and B. pingnanense