Abstract
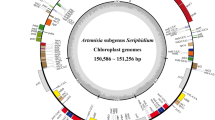
Begonia L. is the only species-rich genus in the Begoniaceae family, comprising more than recorded 2,000 species. However, most of the species in this genus are narrowly distributed, and the phylogenetic relationships of some Begonia sections are still unresolved. Begonia asteropyrifolia Y.M. Shui & W.H. Chen which belongs to the Begonia Sect. Coelocentrum has been listed as an endangered species in China. Due to a shortage of studies on the chloroplast genome of B. asteropyrifolia, studies of comparison and phylogeny with other related species were still scarce. Here, we sequenced the chloroplast genome of B. asteropyrifolia via the Illumina NovaSeq 6000 platform. The complete chloroplast genome of B. asteropyrifolia was 169,512 bp long with a typical circular quadripartite structure, containing a large single-copy (LSC) region (76,310 bp), a small single-copy (SSC) region (18,058 bp) and two inverted repeats (IR) regions (37,572 bp each). The chloroplast genome encoded 113 unique genes, containing 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. The basic structure, gene content and organizations, and number of long repeats in B. asteropyrifolia are similar to previously reported congeneric species. A total of 5,759 (range 144 to 205) SSRs, 1,568 (49 each) long repeats, and 25 highly divergent regions (Pi > 0.03) were detected as candidate molecular markers for further population genetic study and species identification. Moreover, the phylogenetic analysis based on the chloroplast genomes strongly supported the division of 32 Begonia species into six groups related to geographical distribution, and the B. asteropyrifolia, Begonia ferox C.-I Peng & Yan Liu, Begonia gulongshanensis Y.M. Shui & W.H. Chen and Begonia arachnoidea C.-I Peng, Yan Liu & S.M. Ku are closely related and form one clade (Sect. Coelocentrum), which was a subclade in the Asian Clade D of Asian Begonia. Overall, the current study will be beneficial for designing new species-specific DNA markers for new species identification, evolutionary history, and conservation within Begonia.







Similar content being viewed by others
Data availability
The data that support the findings of this study are openly available in the NCBI database.
References
Amiryousefi A, Hyvönen J, Poczai P (2018) IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34(17):3030–3031
Ardi WH, Campos-Domínguez L, Chung KF, Dong WK, Drinkwater E, Fuller D, Gagul J, Garnett GJL, Girmansyah D et al (2022) Resolving phylogenetic and taxonomic conflict in Begonia. Edin J Bot 79(1928):1–28
Beier S, Thiel T, Münch T, Scholz U, Mascher M (2017) MISA-web: A web server for microsatellite prediction. Bioinformatics 33:2583–2585
Chakraborty S, Yengkhom S, Uddin A (2020) Analysis of codon usage bias of chloroplast genes in Oryza species. Planta 252:67
Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK (2006) The complete chloroplast genome sequence of Pelargonium×hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol 23:2175–2190
Clement WL, Tebbitt MC, Forrest LL et al (2004) Phylogenetic position and biogeography of Hillebrandia sandwicensis (Begoniaceae): a rare Hawaiian relict. Am J Bot 91(6):905–917
Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res 45(4):e18
Dong WP, Xu C, Li CH, Sun JH, Zuo YJ, Shi S, Cheng T, Guo JJ, Zhou SL (2015) ycf1 the most promising plastid DNA barcode of land plants. Sci Rep 5:8348
Doyle JJ, Doyle JL (1987) A rapid procedure for DNA purification from small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Forrest LL, Hughes M, Hollingsworth PM (2005) A phylogeny of Begonia using nuclear ribosomal sequence data and morphological characters. Syst Bot 30(3):671–682
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32:273–279
Gao L, Yi X, Yang YX, Su YJ, Wang T (2009) Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: Insights into evolutionary changes in fern chloroplast genomes. BMC Evol Biol 9:130
Guan KY, Yamaguchi H, Li JX, Li HZ, Ma H (2007) Traditional uses of begonias (Begoniaceae) in China. Acta Bot Yunnanica 29:58–66
Guisinger MM, Kuehl JV, Boore JL, Jansen RK (2011) Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol Biol Evol 28:583–600
Ivanova Z, Sablok G, Daskalova E, Zahmanova G, Apostolova E, Yahubyan G, Baev V (2017) Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Front Plant Sci 8:204
Jiao Y, Jia HM, Li XW, Chai ML, Jia HJ, Chen Z, Wang GY, Chai CY et al (2012) Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra). BMC Genomics 13:201
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R (2001) REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29:4633–4642
Li Y, Wang ZH, Xing H, Chi XF, Chen SL, Gao QB (2019) The complete chloroplast genome of Saxifraga sinomontana (Saxifragaceae) and comparative analysis with other Saxifragaceae species. Braz J Bot 42:601–611
Li LF, Chen XL, Fang DM, Dong S, Guo X, Li N, Campos-Dominguez L, Wang W, Liu Y et al (2022) Genomes shed light on the evolution of Begonia, a mega-diverse genus. New Phytol 234:295–310
Li QQ, Chen X, Yang DF, Xia PG (2023) Genetic relationship of Pleione based on the chloroplast genome. Gene 858:147203
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu X, Chang EM, Liu JF, Jiang ZP (2021) Comparative analysis of the complete chloroplast genomes of six white oaks with high ecological amplitude in China. J for Res 5:2203–2218
Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ et al (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18
Moonlight PW, Ardi WH, Padilla LA, Chung KF, Fuller D, Girmansyah D, Hollands R, Jara-Muñoz A et al (2018) Dividing and conquering the fastest-growing genus: towards a natural sectional classification of the mega-diverse genus Begonia (Begoniaceae). Taxon 67(2):267–323
Nguyen LT, Schmidt HA, von Haeseler A et al (2014) IQ-TREE: a fast and efective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Nock CJ, Baten A, King GJ (2014) Complete chloroplast genome of Macadamia integrifolia confirms the position of the Gondwanan early-diverging eudicot family Proteaceae. BMC Genomic 15(Suppl 9):S13
Ogoma CA, Liu J, Stull GW, Wambulwa MC, Oyebanji O, Milne RI, Monro AK, Zhao Y, Li D-Z, Wu Z-Y (2022) Deep insights into the plastome evolution and phylogenetic relationships of the tribe Urticeae (Family Urticaceae). Front Plant Sci 13:870949
Pauwels M, Vekemans X, Godé C, Frérot H, Castric V, Saumitoul-aprade P (2012) Nuclear and chloroplast DNA phylogeography reveals vicariance among European populations of the model species for the study of metal tolerance, Arabidopsis halleri (Brassicaceae). New Phytol 193:916–928
Pombert JF, Lemieux C, Turmel M (2006) The complete chloroplast DNA sequence of the green alga Oltmannsiellopsis viridis reveals a distinctive quadripartite architecture in the chloroplast genome of early diverging ulvophytes. BMC Biol 4(1):3
Qu XJ, Moore MJ, Li DZ, Yi TS (2019) PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15:50
Rajbhandary S (2015) Traditional Uses of Begonia Species (Begoniacae) in Nepal. J Nat Hist Mus 27:25–34
Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM et al (2007) Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics 8:174
Ren T, Yang YC, Zhou T, Liu ZL (2018) Comparative plastid genomes of Primula species: Sequence divergence and phylogenetic relationships. Int J Mol Sci 19(4):1050
Shen J, Zhang X, Jacob BL, Zhang HJ, Deng T, Sun H, Wang HC (2020) Plastome evolution in Dolomiaea (Asteraceae, Cardueae) using phylogenomic and comparative analyses. Front Plant Sci 11:376
Shi C, Liu Y, Huang H, Xia EH, Zhang HB, Gao LZ (2013) Contradiction between plastid gene transcription and function due to complex posttranscriptional splicing: an exemplary study of ycf15 function and evolution in Angiosperms. PLoS ONE 8(3):e59620
Shui YM, Chen WH (2005) New data of sect coelocentrum (Begonia) in Begoniaceae. Acta Bot. Yunnanica 27(4):355–374
Shui YM, Chen WH, Peng H, Huang SH, Liu ZW (2019) Taxonomy of Begonias. Yunnan Science and Technology Press, Kunming
Somaratne Y, Guan DL, Wang WQ, Zhao L, Xu SQ (2019) Complete chloroplast genome sequence of Xanthium sibiricum provides useful DNA barcodes for future species identification and phylogeny. Plant Syst Evol 305:949–960
Tian DK, Xiao Y, Tong Y, Fu NF, Liu QQ, Li C (2018) Diversity and conservation of Chinese wild Begonias. Plant Diversity 40:75–90
Tian SB, Lu PL, Zhang ZH, Wu JQ, Zhang H, Shen HB (2021) Chloroplast genome sequence of Chongming lima bean (Phaseolus lunatus L.) and comparative analyses with other legume chloroplast genomes. BMC Genomics 22:194
Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S (2017) GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res 45:W6–W11
Tseng YH, Hsieh CL, Campos-Domínguez L, Hu AQ, Chang CC, Hsu YT, Kidner CA, Hughes M, Moonlight PW, Hung CH, Wang YC, Wang YT, Liu SH, Girmansyah D, Chung KF (2022) Insights into the evolution of the chloroplast genome and the phylogeny of Begonia. Edin J Bot 79(408):1–32
Wang LY, Wang J, He CY, Zhang JG, Zeng YF (2021) Characterization and comparison of chloroplast genomes from two sympatric Hippophae species (Elaeagnaceae). J Forestry Res 32(1):307–318
Wang NJ, Chen SF, Xie L, Wang L, Feng YY, Lv T, Fang YM, Ding H (2022a) The complete chloroplast genomes of three Hamamelidaceae species: comparative and phylogenetic analyses. Eco and Evol 12:e8637
Wang ZJ, Cai QW, Wang Y, Li MH, Wang CC, Wang ZX, Jiao CY, Xu CC, Wang HY, Zhang ZL (2022b) Comparative analysis of codon bias in the chloroplast genomes of Theaceae species. Front Genet 13:824610
Weng ML, Blazier JC, Govindu M, Jansen RK (2014) Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol 31(3):645–659
Xie YP, Yang GG, Zhang C, Zhang XW, Jiang XF (2023) Comparative analysis of chloroplast genomes of endangered heterostylous species Primula wilsonii and its closely related species. Ecol Evol 13:e9730
Xiong AS, Peng RH, Zhuang J, Gao F, Zhu B, Fu XY, Xue Y, Jin XF, Tian YS, Zhao W, Yao QH (2009) Gene duplication, transfer, and evolution in the chloroplast genome. Biotechnol Adv 27(4):340–347
Xu WB, Xia BS, Li XW (2020) The complete chloroplast genome sequences of five pinnate-leaved Primula species and phylogenetic analyses. Sci Rep 10(1):20782
Yang J, Vázquez L, Chen XD, Li HM, Zhang H, Liu ZL, Zhao GF (2017) Development of chloroplast and nuclear DNA markers for Chinese oaks (Quercus subgenus Quercus) and assessment of their utility as DNA barcodes. Front Plant Sci 8:816
Yang XM, Zhou TT, Su XY, Wang GB, Zhang XH, Guo QR, Cao FL (2021) Structural characterization and comparative analysis of the chloroplast genome of Ginkgo biloba and other gymnosperms. J for Res 32:765–778
Yao XH, Tang P, Li ZZ, Li DW, Liu YF, Huang HW (2015) The first complete chloroplast genome sequences in Actinidiaceae: genome structure and comparative analysis. PLoS ONE 10:e0129347
Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A (2020) Chloroplot: an online program for the versatile plotting of organelle genomes. Front Genet 25(11):576124
Zhu M, Feng PP, Ping JY, Li JY, Su YJ, Wang T (2021) Phylogenetic significance of the characteristics of simple sequence repeats at the genus level based on the complete chloroplast genome sequences of Cyatheaceae. Ecol Evol 00:1–14
Acknowledgements
This work was supported by the Guangxi Natural Science Foundation Program, China [2020GXNSFBA159052], the Guangxi Science and Technology Base and Special Talents Program, China [guike AD20325009] and the National Natural Science Foundation of China [31960083].
Author information
Authors and Affiliations
Contributions
Q.F.L. collected the sample and analyzed the data. W.H.L. provided research idea and supervised the experiments and research documentation. Q.F.L. and W.H.L. wrote the article. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interests
No potential conflict of interest was reported by the author(s).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, Q., Luo, W. Chloroplast genome sequence of Begonia asteropyrifolia and comparative analysis with other related species. Braz. J. Bot 47, 105–117 (2024). https://doi.org/10.1007/s40415-023-00969-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-023-00969-7