Abstract
The taxonomy of all species of Narcissus (Amaryllidaceae), an important horticultural crop, has not been investigated recently. As a new approach, genome size was determined by flow cytometry with propidium iodide from 375 accessions. The somatic nuclear DNA contents (2C) were shown to range from 14 to 38 pg for the diploids. Narcissus assoanus and N. gaditanus are, based on their nuclear DNA content, removed from section Apodanthi and placed in a new section Juncifolii. The different ploidy levels and species involved were entangled for N . “fernandesii” s.l. and a new allotetraploid form is named here. Section Pseudonarcissus was much more heterogeneous in nuclear DNA content than expected. Sixty-five accessions of N. pseudonarcissus possessed, with 23.7 pg, similar amounts of DNA. However, several species from this section were clearly distinctive in nuclear DNA content. It runs from the diploid N. primigenius with 21.7 pg to the also diploid N. nevadensis with 38.2 pg. Also N. abscissus and N. moleroi are with about 26 pg clearly different from N. pseudonarcissus. For the first time, in 11 accessions, hexaploidy was found in N. pseudonarcissus ssp. bicolor. A new section Nevadensis with 30–39 pg of nuclear DNA was split off from the section Pseudonarcissus with now 21–27 pg. A nonoploid N. dubius with 96.3 pg has by far the highest amount of nuclear DNA and can be calculated to have the highest ploidy ever reported in Narcisssus. The total number of Narcissus species was determined as 36, nine more than in Flora Europaea and they were divided up in two subgenera and 11 sections. Flow cytometry is shown to produce easily obtainable and original systematic data that lead to new insights. Genome size or C-value turns out to be one of the most salient features to define the status of the species in the genus Narcissus.
Similar content being viewed by others
Introduction
Since the articles of Fernandes (1966a, b; 1968a, b; 1975), based strongly on cytological investigations, no complete account of all species based on new data has appeared. This seems the more amazing as Narcissus is a horticultural and pharmaceutical important genus. It exhibits the taxonomic difficulties which arise from narrow endemism in the Mediterranean flora and weak reproductive barriers often lead to solitary hybrid specimens (Blanchard 1990; Thompson 2005). Polyploidy contributes significantly to speciation (Fernandes 1968a, b). After doubling chromosome numbers some of these original sterile hybrids occupy large areas and are now recognized as species. Moreover, a high morphological variability makes it doubtful if any Narcissus species can be identified from comparative measurements (Smythies 1973; Blanchard 1990). Also long-established cultivation, hybridization and selection and subsequent escape and naturalization (Webb 1980) have contributed to the taxonomic problems.
The important cytological work of Fernandes (1951; 1968a, b; 1975) has helped greatly to delineate the species, showing the nature of ancient hybrids and in establishing the hybrid nature of taxa formerly regarded as independent species. Fernandes (1951), accepting 22 species, formed the basis of the 27 species of Webb in Flora Europaea (1980). Later Fernandes (1968a, b) accepted 63 species. Based on Fernandes (1968), even 87 species are accepted in “Botanical names in the genus Narcissus” from the International Daffodil Register, updated September (2006). Here, the opinion of Webb is followed (see for a comparison Table 4), but recently described species and new data resulted in 36 accepted species here.
Graham and Barrett (2004) constructed a phylogenetic tree, based on the sequence data of the ndhF and the trnL-F region of the chloroplast genome. Our results are compared to the results of Graham and Barrett, although their investigations were restricted to 36 of the 65 species as recognized by Blanchard (1990). Consequently, several species and subspecies measured here were not included. Moreover, citing Graham and Barrett (2004): “If taxa of hybrid origin are not removed from analysis this can seriously mislead phylogenetic inference”. They removed therefore N. × tortifolius Fern. Casas and N. dubius Gouan from analysis but they were not aware of other ancient hybrids like N. fernandesii Pedro s.l., N. hedraeanthus (Webb & Heldr.) Colmeiro, N. serotinus L. s.l. and may be N. cavanillesii Barra & G. Lopez).
Nuclear DNA content has not often been exploited for taxonomic purposes. It can conveniently be measured by flow cytometry using propidium iodide, a stoichiometric DNA stain that intercalates in the double helix. Where many species in a genus have the same chromosome numbers, differences in nuclear DNA content are often present and have proven to be very effective in delimiting infrageneric divisions in a number of taxa (Ohri 1998). Genome size has been demonstrated to often vary a factor 1.5–3 between taxa with identical chromosome numbers (Ohri 1998; Yokoya et al. 2000; Zonneveld 2001; Zonneveld and van Iren 2001; Ellul et al. 2002; Zonneveld et al. 2003a, b; Hirsch et al. 2004: Albach and Greilhuber 2004; Zonneveld and Duncan 2006). Moreover, Greilhuber (1998, 2005) has clearly shown that intraspecific variation of genome size is much less than assumed. The evolution of genome size (Cx-value, Greilhuber 1979) has received increased attention during recent years (Bennett and Leitch 2005). Primitive angiosperms are now supposed to have had small genomes; increases up to a factor 1,000 have occurred independently in various modern taxa (Leitch et al. 1998). The low intraspecific variation in most Narcissus species shows that genome size can be used as one of the basic factors demarcating infrageneric taxa.
Repetitive DNA elements, including retro-transposons, are major components of eukaryotic genomes and such elements have a tendency towards amplification (Bennetzen and Kellogg 1997; Kalender et al. 2000). Major decreases in genome size occur less frequently and such decreases have been observed especially following a doubling of the total genome by polyploidization (Ohri 1998), or the change from perennial to annual habit (Bennett 1972).
A complete review of the genus Narcissus is long overdue knowing its importance as a horticultural crop. So far genome size was determined in only three species: N. hedraeanthus, N. cantabricus DC. and N. poeticus L. with respectively, 13.1, 14.5 and 27.5 pg (Gonzalez et al. 1990). The relationship between its species is the more important as Narcissus has attracted attention as a source of potentially valuable pharmaceuticals like galanthamine that might have application in the treatment of Alzheimer disease (Colina 2002). In this study, nuclear DNA content was introduced as a novel approach for the study of Narcissus taxonomy. A total of 375 different accessions representing nearly all known species and representatives of 43 natural hybrids were investigated.
Materials and methods
Plant material
Plant material was mainly obtained from the collections of J. Blanchard (UK) and D. Donnison-Morgan (UK), but specimens were also obtained from The Netherlands (C.P. Breed, C. van de Veek, W. Lemmers, Hortus Bulborum Limmen and S. de Groot), from the UK (J.M. Grimshaw, M. Salmon and I. Young), from France (L. de Jager) and from the USA (N. Wilson and H. Koopowitz). Where possible, material of known wild origin was used, and care was taken to ensure correct identification of all material, relying also on the expert opinions of J. Blanchard, D. Donnison-Morgan and S. de Groot. Leaves were used from live plants from the wild or available in the different collections.
Determination of nuclear DNA content
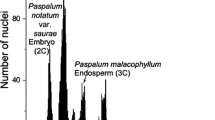
For the isolation of nuclei, about 0.5 cm2 of a fresh full-grown leaf was chopped together with a piece of Agave americana L. or Agave stricta Salm-Dyk as internal standard. The nuclear DNA content (2C-value) of A. americana was measured as 15.9 pg per nucleus with human leukocytes (2C = 7 pg, Tiersch et al. 1989) as the standard. A. stricta with 7.8 pg was used for those sections with DNA values below 20 pg. The chopping was done with a new razor blade in a Petri dish in 0.25 ml nuclei-isolation buffer, with 0.01% RNAse added, as described by Zonneveld and van Iren (2001). After adding 1.8 ml propidium iodide solution (50 mg/l in isolation buffer) the suspension with nuclei was filtered through a 30 μm nylon filter. The fluorescence of the nuclei was measured, 30 and 60 min after addition of propidium iodide, using a PARTEC CA-II flow cytometer. The more DNA is present in a nucleus, the higher is the intensity of the fluorescence. The 2C DNA content of the sample was calculated as the sample peak mean, divided by the Agave peak mean, and multiplied with the amount of DNA of the Agave standard. From chopping of a leaf piece of 0.5 cm2 about 50,000 nuclei could be isolated. For each clone, two to six different runs (determinations) with at least 3,000–5,000 nuclei were measured with two runs from a single nuclear isolation.
Pollen vitality staining
To obtain pollen from plants of section Nevadensis and N. hedraeanthus, anthers were removed just before opening of the flower. Vitality of pollen was determined by suspending and staining the dry pollen into a drop of Cotton Blue Lactophenol (BDH). Empty pollen stays colourless. The percentage of blue pollen grains was considered as an upper value for viability. At least 300 grains were assayed microscopically and in each case three determinations were done.
Results
Based on the work of four consecutive years, nuclear DNA contents of 375 accessions of Narcissus attributed to 36 species in 11 sections are listed in Table 1. Fernandes (1977) remarks: Il serait donc d’intérêt de mesurer la quantité d’acide désoxyribonucléique existent dans les deux groupes (i.e. section Pseudonarcissus and section Ganymedes). This suggestion of the eminent expert on the genus is followed here and extended to all sections. Fernandes (1951) divides Narcissus into two subgenera, subgenus Hermione with the basal chromosome number of n = 5 (11) and subgenus Narcissus with basal chromosome number n = 7 (13). This is followed here, although these two subgenera overlap in nuclear DNA content with for the diploids in subgenus Hermione 20.9–37.4 pg and in subgenus Narcissus 14.2–38.7 pg. This is also in accordance with the phylogeny of Graham and Barrett (2004) based on the sequences of two chloroplast genes. The division here in species and sections, based on DNA amounts, largely follows Webb (1978, 1980), (but not in his treatment of section Jonquillae s.l.) and also takes into account the results of Graham and Barrett (2004). Also a new section Nevadensis is added here. A low intraspecific variation is found in most cases. The interspecific variation shows that genome size in diploid Narcissus varies between 14 pg in the section Bulbocodium to 38 pg in Nevadensis. The difference between the highest and lowest DNA contents is about 24 pg, equivalent to nearly 3 × 1010 bp. This must be the result of a vast number of genomic changes. Even a single picogram difference equates to a difference of 1,000,000,000 bp. Increases in ploidy are accompanied by increases in amount of nuclear DNA. The discussion of species and sections follows the classification presented in Table 1. The allocation to synonymy by Fernandes (1951) and Webb (1978) is largely followed here for the more obscure names. To prevent repeating the same sentence, the picograms used in the discussion below are the average amounts of nuclear DNA for the number of accessions of a certain species as can be found in Table 1. For determination of the standard deviation all relevant measurements were used. Recent hybrids found as solitary plants (in contrast to ancient hybrids that are here treated as species) are presented in Table 2. A resume of the main results compared with those of Webb (1978, 1980) is given in Table 4.
From the data presented it cannot be concluded where or when there was an increase or a decrease or both in the amount of nuclear DNA. The results are compared and discussed in the respective sections. I refrain in most cases to express an opinion on ecological adaptation or geographical distribution in relation to nuclear DNA content as that needs an additional set of data.
Discussion
Genus Narcissus
Subgenus Hermione (Haw.) Spach
Section Serotini Parl.: N. serotinus L., N. miniatus Koop., Donnison-Morgan, Zonn. The rare N. serotinus with 2n = 10 and 20.9 pg is now separated from N. miniatus, widespread along the Mediterranean coasts, with 2n = 30, but with 51.3 pg. This amount of nuclear DNA does not fit a hexaploid N. serotinus, suggested by its chromosome number, as that would have 3 × 20.9 pg = 62.7 pg (Donnison-Morgan et al. 2006). The intermediate nature of the morphology of the ancient hybrid N. miniatus and the amount of nuclear DNA shows that it is actually a doubled form of the hybrid N. serotinus (2n = 10; 20.9 pg) × N. elegans (Haw.) Steud. (2n = 20; 30.2 pg). Knowing the amounts of nuclear DNA this was easy to spot, especially so, as these are all three autumn flowering species. Lifante and Camacho (2007) did extensive morphological measurements on these taxa and concluded that, apart from the two parents a third form, a hybrid, was involved. They did not refer to the earlier work of Donnison-Morgan et al. (2006). Narcissus obsoletus (Haw.) Steud., used subsequently for different taxa by various authors, was equated with this hybrid (Lifante and Camacho 2007). However this application is doubtful as they do not make a distinction between the diploid hybrid N. × obsoletus (N. elegans × N. serotinus) (Table 2) found as solitary specimen amidst its parents in Morocco (Maire 1959) and its widespread tetraploid form N. miniatus (Table 1). Therefore, N. obsoletus is better considered as a nomen confusum or should be confined to the diploid hybrid. Chloroplast DNA data of Graham and Barrett (2004) did not allow them to discriminate between N. serotinus (20.9 pg), N. elegans (30.2 pg) and N. tazetta (30.3 pg). May be the widespread N. miniatus (51.3 pg) was masquerading for N. serotinus.
Section Tazettae DC.: N. broussonetii Lag. y Seg., N. elegans (Haw.) Spach, N. papyraceus Ker Gawl., N. tazetta L. The values for the species of this section vary from 30.2 to 37.4 pg. Narcissus tazetta including the ssp. aureus Baker, range from 30.0 to 31.8 pg with an average of 30.3 pg. Although the chromosome numbers 2n = 20 point to ancient tetraploidy (N. serotinus has 2n = 10), Fernandes (1966a, b; 1968a, b) has shown they behave now as diploids forming only bivalents in meiosis. If N. tazetta is considered as a diploid, five accessions must then be considered as being triploid. They have a 50% higher amount of DNA and include the so-called N. “Chinese Sacred Lily” (2n = 30) that centuries ago must have naturalized in China and Japan. The 16 values for N. papyraceus including the ssp. panizzianus (Parl.) Arcang., ssp. polyanthus (Loisel.) Asch. & Graebn. and ssp. pachybolbus (Durand) Webb have an average of 33.7 pg. The more than 3 pg difference in DNA content between N. tazetta and N. papyraceus supports the separation of these two taxa. It seems likely that authentic N. tazetta has 2n = 20 and N. papyraceus 2n = 22. Rapports on plants with 2n = 21 are then likely based on sterile hybrids (Fernandes 1966b). The three accessions of N. elegans from Morocco have on average 30.2 pg as stated above.
Based on published chromosome numbers (2n = 22, 44), tetraploids (N. broussonetii f. grandiflorus Maire) are found as solitary plants among the diploids (Maire 1959). Only plants with on average 37.6 pg were measured here in six accessions. In flower, in karyotype, in chromosome number and in the amount of nuclear DNA (assuming that the measured plants are all diploids) it resembles N. papyraceus with which it is easy to cross (Maire 1959), so section Aurelia, containing only N. broussonetii, is placed here in section Tazettae. This is also in accordance with the phylogeny of Graham and Barrett (2004).
Subgenus Narcissus
Section Bulbocodii DC.: N. bulbocodium L., N. cantabricus DC., N. hedraeanthus (Webb & Held.) Colmeiro, N. obesus Salisb., N. romieuxii Braun-Blanq. & Maire. Amounts of nuclear DNA in this section range from 14.2 to 15 pg for the diploids and 28.7–47.3 pg for the polyploids. Narcissus bulbocodium has the most numerous and the most different types of polyploids of all Narcissus species (Fernandes 1963). He showed that some populations had triploids and hexaploids but not tetraploid plants. This suggests an origin of the hexaploids from non-reduced gametes. Moreover, the higher the ploidy, the wetter the localities were they were found (Fernandes 1963). For the plants here measured two diploids, six tetraploids, one pentaploid, one hexaploid and one heptaploid was found, the latter two from commerce. Fernandes (1963) reports also on an occasional octoploid that would have 4 × 14.2 = 56.8 pg. Narcissus cantabricus seems the white counterpart of the yellow flowering N. bulbocodium. Contrary to N. bulbocodium, only diploids and tetraploid forms, each with four accessions, were measured. Although the amounts of nuclear DNA are very similar, Fernandes (1959) found that the hybrid between two diploid forms of these species was nearly fully sterile and their karyograms were different. It seems that N. cantabricus originated from N. bulbocodium through structural chromosome changes (Fernandes 1959). Narcissus romieuxii from Morocco was considered by Fernandes (1963) to be the ancient allotetraploid of N. bulbocodium and N. cantabricus. The amount of nuclear DNA of 28.8 pg for all subspecies of N. romieuxii does not counter this. Narcissus bulbocodium, with a nuclear DNA-content of only 14 pg, has by far the lowest amount of DNA of any Narcissus species. This might indicate a plesiomorphic position within the genus, but this is not reflected in the phylogeny based on plastid data by Graham and Barrett (2004). There the Bulbocodium section together with the Pseudonarcissus section formed a well supported clade. With scarcely developed perianth segments, N. bulbocodium s.l. is morphologically the opposite of the other supposed “primitive” species N. cavanillesii with a diminutive corona. Both may be adaptations to a dryer, southerly climate.
Narcissus hedraeanthus, although sometimes included in N. bulbocodium as N. bulbocodium ssp. hedraeanthus (Webb & Heldr.) Voss, is clearly different from N. bulbocodium and N. cantabricus with its 15.0 pg measured in 14 accessions.
The flower shape and the small plant size of N. hedraeanthus reminds of a N. bulbocodium × N. triandrus hybrid. Blanchard (1990) remarks that N. hedraeanthus can be grown from seed but that it carries very small capsules containing far fewer seeds than is usual with the species of this section (apomixis?). Moreover, I measured only 80 and 90% good pollen in two accessions of N. hedraeanthus. As the amount of nuclear DNA is intermediate between the supposed parents, an ancient hybrid origin for N. hedraeanthus cannot be excluded.
Narcissus obesus is a tetraploid but with 2n = 4x = 26 chromosomes instead of the regular 28. As the amount of nuclear DNA is, with 26.5 pg in three accessions, also 2 pg lower than those of a tetraploid N. bulbocodium, it can be calculated that it has lost 7% of its nuclear DNA content. Fernandes (1963) shows that N. obesus is homozygous for a single Robertsonian fusion with loss of the short arms which reduces the chromosome number from 28 to 26.
Section Ganymedes (Haw.) Schult.f.: N. pallidulus Graells, N. lusitanicus Dorda & Fern. Casas, N. triandrus L. Nuclear DNA content in this section varies from 16.9 to 19.0 pg. For the mainly yellow flowering N. lusitanicus from Portugal 16.9 pg is found. The pale yellow N. pallidulus with four accessions from below and around Madrid have a nuclear DNA content of 18.1 pg. The robust, whitish flowering N. triandrus with six accessions from above Madrid up to the Cantabrian Mountains has on average 19.0 pg. The latter includes the isolated N. triandrus “Loiseleurii” or “Capax” from the Iles Glenans, France, considered by Webb (1980) as a separate subspecies. Apart from a different nuclear DNA content they can be separated in plants with a leaf width of about 2 mm for N. pallidulus and N. lusitanicus and more than 4 mm for N. triandrus (Blanchard 1990). Narcissus triandrus is usually divided in three varieties or subspecies (Webb 1980; Barra Lazaro 2000). However Dorda and Fernandez Casas (1989) considered them to be separate species and also Perez-Barrales et al. (2006) did so, based on chloroplast DNA data. Our data support three species, although it cannot be excluded that the presence of B-chromosomes might influence this difference. Blanchard (1990) did find all three colours in single populations. However, it cannot be excluded that each species occasionally can show all three colours or that they occasionally form hybrid swarms.
Section Juncifolii (A. Fern.) Zonn. sect nov. Typus N. assoanus Dufour Other species: N. gaditanus Boiss. & Reuter).
Based on subsection Juncifolii A. Fern. Bol. Soc. Brot.II, 40: 207–261 (1966).
The rare N. gaditanus Boiss. & Reuter confined to southern Spain and Portugal, has smaller but more numerous flowers to a scape compared to the widespread N. assoanus from northern Spain and southern France. Narcissus gaditanus and N. assoanus are closely related with 18.8 and 19.3 pg, respectively, and are here separated from section Jonquillae with about 33 pg for the diploid forms. This corroborates with the plastid DNA data of Graham and Barrett (2004) where they are also placed separately from the Jonquillae clade. A strong cytological difference was already observed by Fernandes (1975) who placed them in a subsection Juncifolii of section Jonquillae. Their unique amount of nuclear DNA is otherwise only found in the unrelated N. triandrus. Because there is only a small difference in morphology and genome size, N. gaditanus is here doubtfully maintained as a species separate from N. assoanus.
Section Tapeinanthes (Herb.) Traub: N. cavanillesii Barra & G. Lopez). Six accessions of N. cavanillesii were measured, from Spain as well as from Morocco with an average value of 33.3 pg. Hardly any difference was found between these localities. Chromosome numbers are reported as 2n = 4x = 28 (Fernandes and Fernandes 1951). The species most similar, excluding N. bulbocodium and N. triandrus s.l. on morphological grounds, is N. assoanus/gaditanus of section Juncifolii with about 19 pg. Fernandes (1975) remarks on the analogy between the karyotypes of N. cavanillesii and N. assoanus. If N. cavanillesii is an ancient allotetraploid, the amount of nuclear DNA suggests that N. assoanus (including N. gaditanus) and N. bulbocodium could be the parents. Graham and Barrett (2004) placed them all three in the same section with N. pseudonarcissus in their phylogenetic tree. However, the nearly absent corona in N. cavanillesii seems to contradict this. A single plant from the Chelsea Physics Garden, England had 17.7 pg. So if properly diagnosed it could be a diploid form of N. cavanillesii. This species is considered to be a primitive narcissus with hardly a corona and was until recently even placed in a separate genus, Tapeinanthus Herb.
Section Apodanthi A. Fern.: N. calcicola Mendonsa, N. rupicola Dufour ex Schult.f., N. scaberulus Henriq. The species of section Apodanthi with about 26 pg are very similar in this respect and have very similar karyotypes (Fernandes 1975). They differ from section Jonquillae also by their karyotypes and the presence of strophioles. Narcissus scaberulus with scabrid leaves and an incurved corona is considered here as a distinct species despite a similar amount of DNA as was found in N. calcicola. The results of Graham and Barrett (2004) also show a well supported split between N. calcicola on one side and N. albimarginatus D. & U. Mull.-Doblies, N. atlanticus Stern, N. cuatrecasasii Fern. Casas, Lainz & Ruiz Rejon, N. rupicola and N. scaberulus, on the other side. Narcissus watieri Maire and N. marvieri Jahand. & Maire both from Morocco are considered here as subspecies of the Spanish N. rupicola. This is in accordance with the results of Perez-Barrales et al. (2006) based on chloroplast DNA sequences. The similarity in average DNA content may infer close phylogenetic relationships between these members of section Apodanthi. They are all morphologically very similar but differ in that N. rupicola s.l. has only a single flower per stem and N. calcicola and N. scaberulus have several. Narcissus cuatrecasasii was removed from section Apodanthi to section Jonquillae (see below).
Section Jonquillae DC.: N. blanchardii Zonn., N. cuatrecasasii Fern. Casas, Lainz & Ruiz Rejon, N. jonquilla L., N. viridiflorus Schousb. Webb (1980) included subsection Juncifolii and section Apodanthi here. These are here treated as separate sections based on their vastly different amounts of nuclear DNA. The diploid species in this section have similar amounts of nuclear DNA with 31.7 and 32.9 pg. However all kinds of polyploids are found in this section too. To start with the easy one: N. viridiflorus is an autumn flowering narcissus with unique green flowers. It is a tetraploid (2n = 4x = 28) and has an average of 63.5 pg DNA. It clearly fits this section as a N. jonquilla type with a doubled nuclear DNA content. Narcissus jonquilla ssp. jonquilla has 32.8 pg for seven accessions. Narcissus cordubensis Fern. Casas with 33.2 pg is here treated as a subspecies of N. jonquilla.
Analyses of morphological characters (Webb 1978) and plastid DNA sequences (Graham and Barrett 2004) place N. cuatrecasasii in section Apodanthi. The amount of nuclear DNA however, 31.7 pg, based on four accessions, is clearly different from the about 26 pg of DNA of the remaining species in that section. Fernandes (1968a) also states that the karyotype of N. cuatrecasasii is different from the other species of section Apodanthi there. It is provisionally placed in section Jonquillae here and may be it is even only a form of wet mountains and should be named N. jonquilla ssp. cuatracasasii.
The difficult part was N. fernandesii where opinions differ and results were at first difficult to interpret. Webb (1978) leaves the species as incertae sedis and remarks that it is in most characters intermediate between N. (jonquilla ssp.) willkommii (Samp.) A. Fern. and N. gaditanus. Fernandez Casas (1996) considers them all, from diploids up to hexaploids, as forms of the ancient hybrid N. jonquilla × N. assoanus and that is not far away from what is proposed here. He places all these forms under N. fernandesii. When more accessions were measured, the following solution presented itself. Five diploid accessions of N. “fernandesii” were found with 33.0 pg. These are here considered as N. jonquilla ssp. fernandesii, as was already suggested by Fernandes (1966a). Fernandes (1968a) remarks that the diploid N. fernandesii could not be found anymore in the locus classicus in Portugal, but later several populations were located in Spain. However the bulbs he obtained from Gomos Pedro, the author of this species, had 2n = 14. The most common in nature (Fernandes 1966a) were the N. “fernandesii” with 2n = 28, that are found here to have 51.2 pg DNA. Fernandes remarks that it has two different sets of chromosomes that form bivalents only and are fully fertile. However, he considers it despite these results as autotetraploids and named it as N. fernandesii var. major. Based on nuclear DNA amounts, these cannot be autotetraploids of N. (jonquilla ssp.) fernandesii. The amount of nuclear DNA clearly points to the fact that it must be an allotetraploid as is shown below.
Narcissus blanchardii Zonn. stat. nov.
Ancient allotetraploid form of N. jonquilla (ssp. fernandesii) × N. gaditanus
Based on N. fernandesii var. major A. Fern. in: Bol. Soc. Brot. II, 40: 239(1966). Named after John Blanchard who wrote an eminent book on Narcissus.
The most difficult to interpret at first were the three samples of N. “fernandesii” (Table 2) with 42.5 pg that cannot be triploid forms of N. jonquilla ssp. fernandesii as these would have 1.5 × 32.8 = 49.2 pg. Fernandes (1966a) describes diploids of N. willkommii with 2n = 14 and considers the plant with 2n = 21 as an allotriploid. The diploid N. willkommii is considered by Fernandes (1966a) to be derived from N. jonquilla. It has a fitting 33.3 pg of nuclear DNA and is here considered as N. jonquilla ssp. willkommii (A. Fern) Zonn. It was already placed under N. jonquilla by Coutinho (1939). The triploid form of N. “fernandesii” that was measured gave a fitting DNA value for the hybrid N. jonquilla ssp. fernandesii (Pedro) Zonn. (two sets of chromosomes) and N. gaditanus (one set of chromosomes): 33.8 + 9.4 = 43.2 pg. It seems related to the triploid described by Fernandes (1966a) as N. willkommii (non-reduced) × N. gaditanus. Montserrat and Vives (1991) even found a hexaploid form of N. fernandesii and this could be the doubled form of the allotriploid mentioned here.
If it is supposed that all N. fernandesii s.l. (excluding the diploids that are forms of N. jonquilla) are hybrids of N. jonquilla × N. gaditanus it must be remarked that we did not encounter the diploid, or the pentaploid or the hexaploid hybrid forms. A plant from the same locality from where the hexaploid hybrid was reported (Santuario de la Virgen de la Cabeza) had a diploid amount of DNA and is considered to be N. jonquilla ssp. fernandesii.
Section Pseudonarcissus DC.: N. abscissus (Haw.) Roem. & Schult., N. asturiensis (Jord.) Pugsley, N. cyclamineus DC., N. moleroi Fern. Casas, N. primigenius (Fern. Suarez ex Lainz) Fern. Casas & Lainz, N. pseudonarcissus L. This section is characterized, apart from its large corona, by its uniseriate anthers, biseriate in the other sections (Blanchard 1990). A large number of species has been described in this section. This has surely to do with the fact that these species are the most popular spring flowers. Numerous hybrids are found and named in the wild and thousands of cultivars are raised and do occasionally escape. Some authors considered that there is just a single variable species in this section, like Burbridge and Baker (1875) or Rouy (1912), others like Pugsley (1933) came up with 27 species, eight of which are accepted here. Since 1933 16 more species were described in this same section of which only three are accepted here (Table 1). Webb (1980) is mainly followed here. Those species that have very similar amounts of DNA, are here all considered to be at most subspecies if not synonyms. Narcissus alpestris Pugsley, N. cernuus Salisb., N. confusus Pugsley, N. lobularis Schult. f., N. nanus (Haw.) Spach, N. perez-chiscanoi Fern. Casas, N. provincialis Pugsley, N. pumilus Salisb. and N. radinganorum Fern. Casas are here all considered synonyms of N. pseudonarcissus, for details see Table 1. Eight subspecies are recognized here. Compared with Webb (1980), N. nevadensis is here reinstated as a distinct species and placed in a different section, N. portensis Pugsley as incertae sedis and N. pseudonarcissus ssp. bicolor (L.) Baker, ssp. eugeniae (Fern. Casas) Fern. Casas, and spp. minor (L.) Baker are added here. Narcissus pseudonarcissus ssp. pseudonarcissus (with six accessions), ssp. major (16), ssp. pallidiflorus (2), ssp. eugeniae (4), ssp. moschatus (8) and ssp. minor (8) have on average about 23.7 pg of DNA.
Two accessions of N. primigenius (Fernandez Suarez ex Lainz) Fern. Casas & Lainz (2n = 14, Fernandes 1991) from Riano, northern Spain, had only 21.7 pg. This value is the lowest found in section Pseudonarcissus. This could be indicative of a primitive position, but there are no other arguments. The plant is only half the size of the tetraploid N. pseudonarcissus ssp. nobilis (Haw.) A. Fern and is suggested to be the diploid form of it (Sanudo 1984). Although they are similarly bicoloured, the nuclear DNA content points to independent species. N. asturiensis (eight accessions) and its tetraploid form ssp. villarvildensis (T. E. Diaz & Fern. Prieto) Rivas Mart., T. E. Diaz, Fern. Prieto, Loidi & Penas have an amount of nuclear DNA of 24.2 and 48.2 pg, respectively. They differ from N. pseudonarcissus in their small size and this small difference in amount of DNA. Fernandez and Fernandez Casado (1991) report that N. (pseudonarcissus ssp.) pallidiflorus × N. asturiensis has only 6% good pollen and this might further indicate that they are separate species.
Barra and Lopez (1995) have two subspecies under N. minor L.: ssp. minor (with N. jacetanus Fern. Casas as a synonym and with var. brevicoronatus (Pugsley) Barra & G. Lopez with N. vasconius (Fern. Casas) Fern. Casas as synonym) and ssp. asturiensis (with the vars. villarvildensis T. S. Diaz & Fern Prieto and cuneiflorus Willk. & Lange). On the other hand, Uribe-Echebarria Diaz (1998) divided N. asturiensis in three subspecies: ssp. asturiensis (with ssp. villarvildensis as synonym), ssp. brevicoronatus (Pugsley) Uribe-Ech. and ssp. jacetanus (Fern. Casas) Uribe-Ech. (with N. vasconius as a synonym). Narcissus jacetanus is growing on limestone whereas N. asturiensis grows on acid soils. It is considered to be a separate species here as with only 22.3 pg it seems to differ in this respect also. Moreover, the hybrid N. jacetanus × N. alpestris (=N. pseudonarcissus ssp. moschatus (L.) Baker has only 5% good pollen (Fernandez Casas 1986b).
Four (sub)species are accepted here for these small flowered daffodils: N. pseudonarcissus ssp. minor (23.6 pg), N. primigenius (21.5 pg), N. asturiensis (24.2 and 48.2 pg) and N. jacetanus (22.3 pg). It cannot be excluded that the N. pseudonarcissus ssp. minor measured here), as they all came from cultivated material, including the plant from Linnaeus, are just small forms of N. pseudonarcissus. Based on the difference in nuclear DNA content, N. minor cannot be equated with N. jacetanus as did Barra and Lopez (1995).
There are a few more notable exceptions with respect to nuclear DNA content in the section Pseudonarcissus. Narcissus cyclamineus, long recognized as being morphological different because of its reflexed perianth segments, has 26.0 pg. Graham and Barrett (2004) could not find a difference in plastid DNA sequence between N. cyclamineus and N. asturiensis.
A similar high amount, 26.2 pg, is found for N. abscissus (Haw.) Schult.f. from the Pyrenees. A comparable amount, 26.0 pg, is found in N. poeticus L. Narcissus abscissus is morphologically very similar and has even been equated (Webb 1980) with N. bicolor L., the latter described from a cultivated plant. The close relationship between N. bicolor and N. abscissus suggested by Webb (1980) and others is not confirmed here, as there are clearly differences in ploidy and basic amounts of DNA between these species. Apart from its wild provenance, the main morphological characteristics of N. abscissus, compared to N. bicolor, are a much smaller bulb size, leaves nearly half the width and a cylindrical, seemingly cut-off corona. The corona is sometimes coloured an almost orange yellow (Meadows 1972). However, there is a difference in opinion on the shape of the corona. Whereas Blanchard (1998), Meadows (1972) and Mathew (2002) consistently describe N. abscissus with a truncated corona, Fernandez Casas (1991) and Susanna and Garnatje (2005) describe and figure it as having an expanded one. The latter even writes: “A distinctive trait of N. abscissus… is the intense yellow corona with the margin strongly curved outwards in contrast with the straight pale yellow corona of, e.g. N. pallidiflorus”. The English opinion is followed here as already Haworth (1831) placed N. abscissus and four other species, based on its “clipt-trunke” corona, in a separate genus Oileus (Haw.) (Pugsley 1933).
Another question is whether N. abscissus is a species or a hybrid. It could be speculated that N. abscissus is an (ancient) hybrid, derived from N. poeticus × N. pseudonarcissus. All three taxa can still be found together in de Val d’Aran, Pyrenees (Meadows 1972) and many other places. The F1 hybrid shows a shorter, but also truncated corona. Repeated (back) crosses could have selected the truncated corona and nuclear amount of DNA from N. poeticus, but the length of the corona and its overall appearance from N. pseudonarcissus. Also the almost white to medium yellow tepals, the long pedicels and late flowering might be due to the influence of N. poeticus. Already Engleheart (1894) reported that among the selfed progeny of N. × bernardii DC. ex Henon (N. poeticus × N. pseudonarcissus), forms were found that covered the whole range of variation from N. pseudonarcissus, via N. abscissus to N. × bernardii and N. poeticus (Wylie 1952). I am inclined to consider N. abscissus an ancient hybrid.
A third plant, with 26.1 pg also deviating in the amount of nuclear DNA from N. pseudonarcissus is N. moleroi Fern. Casas from N.E. Spain (Gerona, Barcelona). It is usual considered to be a pale yellow form of the icy-white N. alpestris, but the N. alpestris measured from Cerler-Benasque had only 23.4 pg of nuclear DNA. Narcissus alpestris is a synonym of N. moschatus L., now N. pseudonarcissus ssp. moschatus (L.) Baker. Narcissus moleroi too has a drooping flower, but with a straight pale yellow corona and nearly white petals. Maybe it is a colour form of N. abscissus with a similar amount of DNA. Peculiar is that from the same two places, Col de la Crueta and Cerler, Blanchard reported white flowering narcissi whereas Norman and Norman (2003) report oxlip yellow narcissi. Maybe there is a difference in flowering time (and amount of nuclear DNA!) between the white flowering N. alpestris and the yellowish flowering N. moleroi.
Section Nevadensis Zonn. sect. nov.: N. nevadensis Pugsley. J. Roy. Hort. Soc. 58:17–93 (1933) Scapus multiflores, corona and spatha longissima, DNA contentus nucleorum 30–39 pg a sectione Pseudonarcissus differt. Species: N. bujei Fern. Casas, N. nevadensis, N. longispathus ssp. longispathus, N. longispathus ssp. alcarensis, N. longispathus ssp. segurensis and N. longispathus ssp. yepesii.
Very surprising is the result for the species from just northeast of the Sierra Nevada, N. longispathus, N. alcarensis Rios et al. N. segurensis Rios et al. and N. yepesii Rios et al. (Rios-Ruiz et al. 1998) as these have all 36–38 pg of DNA. These species often have more than one flower to a scape, not found elsewhere in section Pseudonarcissus. The high amount of nuclear DNA seems at first glance easily explained as all four being triploid forms of N. pseudonarcissus (23.7 pg). Although only the chromosome number for N. nevadensis and N. longispathus with similar amounts of DNA is known (2n = 2x = 14, Fernandes 1968; Sanudo 1984), it makes it feasible that the same number will be found in the other three taxa. This means that they have the highest amount of DNA of all diploid Narcissi. For further assurance, the fertility of the pollen was also tested. Having found more than 95% good pollen in N. alcarensis, N. segurensis and N. yepesii, it is highly unlikely that they are triploids. The latter three are here treated therefore as subspecies of N. longispathus. Narcissus nevadensis from the same area has consistently a 2 pg higher amount of nuclear DNA. Also Graham and Barrett (2004) could separate N. longispathus and N. nevadensis with a bootstrap value of 76% and both were well separated from N. pseudonarcissus. Fernandes (1951) considers N. nevadensis to be the most primitive of the section Pseudonarcissus and the Sierra Nevada as the centre of origin of the section. Webb (1980) and others consider it a subspecies of N. pseudonarcissus but with a 12 pg difference that is highly unlikely. With its high amount of nuclear DNA it would then suggest that the other species of this section have lost 50% or so of their DNA without a change in chromosome number, an unlikely event. Graham and Barrett (2004) place N. assoanus and N. gaditanus with 18–19 pg in the same clade as N. longispathus and N. nevadensis with 36–38 pg, although with a low bootstrap value of 56. It seems attractive to suggest an ancient tetraploidy for N. longispathus/nevadensis from N. assoanus/gaditanus. This would fit the amounts of nuclear DNA but would suggest that N. longispathus/nevadensis retained the full amount of DNA and yet at the same time reduced the number of chromosomes again from 2n = 28 to 2n = 14, an unlikely event. Both N. nevadensis and N. longispathus are here kept separate.
Clearly separate from N. pseudonarcissus in nuclear DNA content but hardly morphologically is the plain yellow N. bujei with 30.0 pg, described from the Serrania de Ronda, southern Spain. This value more or less fits N. longispathus (36 pg) × N. spec. with 26 pg. Several populations of section Pseudonarcissus are now found with about 26 pg of DNA. So it comes as no surprise that were they meet plants of section Nevadensis, hybrids like N. bujei are found. This (ancient) hybridisation seems not too far fetched as N. bujei was first described as a variety of N. longispathus (N. longispathus var. bujei Fern. Casas), later as a variety of N. hispanicus and then as a subspecies of it and finally as a species, all by the same author. Fernandez Casas (1986a) reports on seeds in N. bujei having a black strophiole, so it seems to produce seed and its fertility is confirmed by Blanchard (personal communication).
Despite the fact that a large number of accessions of N. pseudonarcissus had similar amounts of DNA of approximately 23.7 pg, several species from this section, as shown above, are clearly distinctive in nuclear DNA content. It runs from the diploid N. primigenius with 21.7 pg to the also diploid N. nevadensis with 38.2 pg. This is contrary to what is found in other sections were most species have a similar amount of DNA. The evidence presented here indicates that comparisons may be based on superficial characters. There are only small differences in the amount of nuclear DNA of the species in the other sections. Therefore, to stay in line with this, the section Pseudonarcissus is divided in two separate sections Pseudonarcissus s.s. with 21–27 pg for the plants from mainly France, northern Spain and Portugal, and Nevadensis with 30–39 pg for the plants from southern Spain.
Section Narcissus: N. poeticus L. Hardly a difference in DNA content was found between N. poeticus and N. radiiflorus, with 26.0 and 26.1 pg. These data strengthen the viewpoint (Webb 1980) based on morphological evidence, that N. radiiflorus cannot be upheld as a separate species. So the latter is indeed best treated as a subspecies: N. poeticus ssp. radiiflorus (Salisb.) Baker. Apart from the 16 diploid accessions, five plants with a triploid amount of DNA and one plant with a tetraploid amount of DNA was found, the latter from cultivated plants. It seems possible that the triploids N. poeticus “Hellenicus” and N. poeticus “Recurvus” described from Greece and Switzerland are actually garden escapes or alternatively were selected from nature for their triploid vigor. Fernandes (1968a) remarks on the similarity of the karyotypes of this section and section Pseudonarcissus. Also Graham and Barrett (2004) place N. poeticus in the Pseudonarcissus clade. As the hybrid between these two seems to be the only fertile intersectional hybrid it is mainly the deviating morphology of the flower, with a very short and orange coloured corona, that has prevented so far to unite these two sections.
Polyploids in Section Pseudonarcissus
The polyploids in this section are here treated separately. Fernandes (1991) found several populations in Spain and Portugal with tetraploid [as N. c.f nobilis (Haw.) Schult. f.] or hexaploid (as N. leonensis Pugsley) chromosome numbers. Sanudo (1984) reported on tetraploid and hexaploid N. nobilis (the latter = N. leonensis?). Based on amounts of nuclear DNA, N. asturiensis ssp. villarvildensis, including the very early flowering N. “Cedric Morris” from Ribalden, Costa Verde and a single accession of N. pseudonarcissus ssp. nobilis were found here to have a tetraploid amount of DNA. Although received under different names 11 hexaploid accessions from four different locations are arranged here under N. pseudonarcissus ssp. bicolor. As expected from its higher ploidy, it has larger bulbs and leaves compared to the often equated diploid N. abscissus. N. pseudonarcissus ssp. bicolor differs in turn by its smaller size and earlier flowering from the very large hexaploid N. pseudonarcissus ssp. leonensis (Fernandes 1991; Table 1). These wild hexaploids are interesting as hardly any are found among the cultivated forms investigated. This seems to exclude that they are escapes from cultivation. Moreover, Wylie (1952) remarked that the optimum level of ploidy in Narcissus cultivars seems to be the tetraploid one. Maybe the hexaploids are discarded in culture as too slow growing.
Sanudo (1984) found a diploid chromosome number for his “N. bicolor” but this could point to the often equated N. abscissus. On the other hand, the N. pseudonarcissus used by Graham and Barrett (2004) has the same accession number (Blanchard 9401) as the plant here determined as the hexaploid N. bicolor. Fernandes and Fernandes (1946) found triploid and tetraploid chromosome numbers for N. bicolor. These lower chromosome numbers are difficult to reconcile with the hexaploid amount of nuclear DNA found in plants here ascribed to N. bicolor. No one so far has reported on hexaploid chromosome numbers in N. bicolor or in N. radinganorum for that matter. The single tetraploid N. nobilis reported here is of doubtful wild origin (Donnison-Morgan, personal communication). An alternative could be to combine all hexaploids (18 measured here) under N. pseudonarcissus ssp. bicolor.
In seven species of Narcissus, N. asturiensis, N. bulbocodium, N. broussonetii, N. cantabricus, N. poeticus, N. pseudonarcissus and N. tazetta, apart from the diploid forms, polyploids are found. Some are considered separate species, some are given separate subspecific names and in some cases there is not a separate name at all. It seems better to give them all a subspecific status when they form separate populations. So instead of the hexaploid N. bicolor and N. leonensis and the tetraploid N. nobilis it seems better to use N. pseudonarcissus ssp. bicolor (L.) Baker, N. pseudonarcissus ssp. leonensis Pugsley and N. pseudonarcissus ssp. nobilis (Haw.) A. Fern. If they are single plants or from cultivated origin like the triploid N. tazetta or the triploid and tetraploid N. poeticus it is better to give them a cultivar name.
Hybrids between species
Forty-three putative primary hybrids of wild origin were also investigated (Table 2). A distinction is made here between ancient hybrids growing over a larger area and therefore considered as species, included in Table 1 and more recent hybrids growing as solitary plants amidst their parents (Table 2). A suggested pairing can be confirmed as probable, when this is between species with distinct DNA values and the expected intermediate DNA value of putative hybrids is readily apparent. Of the 43 hybrids between species here measured, nearly all have an amount of nuclear DNA that is compatible with this calculation. Everywhere were two species of Narcissus meet it seems possible to find the hybrid and most of these have been given names. The occurrence of these hybrids in nature seems a major mechanism for the formation of new species and several instances are indicated above.
Narcissus portensis Pugsley is known from herbarium material only. It is said to be common (!) in central Spain and can be found in several floras, both as a species or as subspecies of N. pseudonarcissus. Unfortunately, not a single plant could be located for measurements. Based on its description, I suspect that it is a hybrid of N. bulbocodium × N. pseudonarcissus. It is clearly very rare in culture and nature. It has, according to the literature, narrow (6 mm) leaves, narrow perianth segments, shorter than the funnel shaped corona, biseriate? anthers and “the flowers give it something of the aspect of a N. bulbocodium” (Pugsley 1933; Gadeceau in Pugsley 1933). Moreover, Fernandez Casas (1986a) speaks of an intersectional hybrid N. × portensis, unfortunately without giving any further details. I suppose he means the same plant.
Interesting is also N. dubius, an allohexaploid plant with 2n = 6x = 50 originating from a doubled hybrid of N. papyraceus × N. assoanus (non-reduced) (Table 3). Eight accessions were measured with an average of 66.1 pg. Most remarkable was the find of a plant with a nonoploid amount of DNA with 96.3 pg. It not only has by far the highest amount of nuclear DNA ever reported in Narcisssus, but it can be calculated to have by far the highest ploidy.
In N. × tortifolius forms with a triploid and tetraploid (2n = 36) amount of DNA are found. Fernandez Casas (1977) reported 2n = 22. This is probably a mistake and more likely should be 2n = 25 for the triploid. The putative parents according to him are N. gaditanus and (N. papyraceus ssp.) panizzianus. The tetraploid (2n = 36, Romero et al. 1983) is then a doubled N. gaditanus × N. panizzianus and the triploid form is then N. gaditanus (non-reduced) × N. panizzianus. Leaving aside the subtle differences between N. papyraceus versus N. panizzianus and N. gaditanus versus N. assoanus, we then have the triploid N. × tortifolius which by doubling its DNA gives the hexaploid N. dubius and from this the nonoploid N. dubius. Furthermore, we have the tetraploid form N. × tortifolius, presumably from the diploid hybrid not encountered here. However, N. koshinomurae Fern. Casas is described (1996) as N. fernandesii × N. panizzianus (in description: Forma externa Narcisso dubio simile, et cum illo interdum confusus!). Blanchard (1998) however, found hybrids in the same population and considers them to be N. assoanus × N. panizzianus. The nuclear DNA content fits the parents suggested by Blanchard, not those by Fernandez Casas. Graham and Barrett (2004) place N. × tortifolius and N. dubius amidst their Jonquillae clade. This is incongruent with the fact that the chloroplast DNA must be of one of the supposed parents either N. panizzianus or N. assoanus, both not in the Jonquillae clade. The data on this N. dubius complex are summarized in Table 3
.
Allopolyploids like N. dubius or N. miniatus bring up the question when a hybrid must be considered as a species. In this article, if not found as single plants but forming one or more populations they are considered to be distinct species. It needs further investigation, e.g. sequencing of nuclear DNA, whether or not plants that are suggested here as possible of hybrid origin, i.e. N. abscissus, N. blanchardii, N. bujei, N. hedraeanthus and N. portensis are indeed (ancient) hybrids.
Two N. bulbocodium look-alike plants had a peculiar amount of DNA. From Mazagon, Andalucia, H. Berkhout did give me a leaf of a single plant with 17 pg and from Cuidad Real, Mestanza there is plant, received as N × barrae Fern. Casas, that is supposed to be N. cantabricum × N. bulbocodium, but has 17.1 pg. If not aneuploids, these two may be considered as N. gaditanus (19.2 pg) × N. bulbocodium (14 pg) as that would fit the amount of nuclear DNA. A N. bulbocodium look-alike plant was also reported by Graham and Barrett (2004) but ended up in their Jonquifoliae clade, suggesting it had a different hybrid origin.
B-chromosomes
Fernandes and Franca (1974) report on B-chromosomes in N. assoanus, N. asturiensis, N. bulbocodium, N. gaditanus and N. hispanicus. Brandham and Kirton (1985) found them in N. assoanus, N. asturiensis, N. bicolor, N. calcicola, N. cyclamineus, N. eugeniae, N. minor, N. tazetta and in N. triandrus. In N. bulbocodium Fernandes (1963) found 0–5 B-chromosomes. They are about two third of the length of the smallest chromosomes and may or may not be heterochromatic. They seem to be widespread and it cannot be excluded that they contribute a few percent to the total amount of DNA as measured here. At first sight they could explain the “anomalous” pseudonarcissi with 26 pg or the differences between the three species in section Ganymedes. However, Fernandez Casas (1996) did not find them in N. cyclamineus or N. moleroi with 26 pg, although he did report on two B-chromosomes in N. eugeniae with 23.2 pg. B-chromosomes might increase the amount of nuclear DNA and will consequently increase nuclear size, cell size and perhaps even plant size. This will lead to a later flowering according Fernandes (1968a). However, it cannot be excluded that B-chromosomes are leftovers from deletions of repeats in the regular chromosomes. In that case the total amount of nuclear DNA will not change at first. Also variation in constitutive heterochromatin can result in intraspecific genome size variation (Greilhuber and Speta 1978; Greilhuber 1995). It would be worthwhile to investigate the relationship between the total amount of nuclear DNA, the number of B-chromosomes and/or amount of heterochromatin present and their effect on any other character.
Conclusions
In Table 4 all Narcissus species have been arranged in two subgenera and 11 sections and compared to the work of Webb (1978, 1980). A strong correlation is found between genome size and the division into sections. Therefore, those few were the amount of nuclear DNA did not fit with other species in the section are here confidently placed in others. The number of 36 species proposed here can be compared to the 26 (+1) species suggested in Flora Europaea by Webb (1980), the 28 species described in the R.H.S. Dictionary of Gardening (1992) or the 22 species of Fernandes (1951) but not with the number of the about 60 species suggested by Fernandes (1968a, b), Nutt (1993) or Blanchard (1990). Especially section Pseudonarcissus turns out to be heterogeneous. It runs from the diploid N. primigenius with 21.7 pg via N. pseudonarcissus with 23.7 pg and N. abscissus with 26 pg to the also diploid N. nevadensis with 38.2 pg. Therefore a new section Nevadensis is introduced. Based on the same arguments section Jonquillae (according to Webb 1978) is now divided in three sections: section Jonquillae s.str. with about 31 p., section Apodanthi with about 26 p. and section Juncifolii with about 19 p. The N. fernandesii complex is now entangled and this has lead to a new taxon N. blanchardii.
Flow cytometry is shown here to be a useful tool to indicate or decide on the taxonomic status of different accessions of Narcissus. This is especially clear if one looks at the last column in Table 1 (“received as”). This method also indicates new and published chromosome counts through the amounts of nuclear DNA. Also several new instances of polyploidy were observed. At the same time the results points out taxonomic problems that need further investigation. Narcissus dubius and N. fernandesii show where even sequencing would not have given the detailed results obtained here with flow cytometry. The speed and cost effectiveness of measuring nuclear DNA content and its predicative accuracy makes it a useful tool for identifying species.
References
Albach DC, Greilhuber J (2004) Genome size variation and evolution in Veronica. Ann Bot 94:897–911
Barra Lazaro A (2000) Dos nuevas variedades de Narcissus triandrus L. Anales Jard Bot Madrid 58:184–186
Barra Lazaro A, Lopez Gonzales G (1995) Sobre la identidad de Narcissus minor L. y plantas afines. Anales Jard Bot Madrid 52:171–178
Bennett MD (1972) Nuclear DNA content and minimum generation time in herbaceous plants. Proc Roy Soc Lond 181:109–135
Bennetzen JL, Kellogg EA (1997) Do plants have a one way ticket to genomic obesity? Pl Cell 9:1509–1514
Bennett MD, Leitch IJ (2005) Nuclear DNA amounts in Angiosperms: progress, problems and prospects. Ann Bot 94:45–90
Blanchard J (1990) Narcissus, a guide to wild daffodils. Published by AGS, Lye End Link, Woking, UK
Blanchard J (1998) Narcissus update. Bull Alp Garden Soc 66:330–337
Brandham PE, Kirton PR (1985) The chromosomes of species, hybrids and cultivars of Narcissus L. (Amaryllidaceae). Kew Bull 42:65–102
Burbridge FW, Baker JG (1875) The Narcissus. London
Colina C (2002) The production of galanthamine by Narcissus tissues in vitro in: Narcissus and daffodils. In: Hanks GR (ed) Narcissus. Taylor & Francis, UK
Coutinho AXP (1939) Flora de Portugal ed. 2, Lisboa
Donnison-Morgan D, Koopowitz H, Zonneveld BJM, Howe M (2006) Narcissus miniatus Donnison-Morgan, Koopowitz & Zonneveld sp. nov. A new species of Narcissus (Amaryllidaceae) from southern Spain. RHS Daffodil, Snowdrop and Tulip yearbook 2005/2006:19–25
Dorda E, Fernandez Casas J (1989) Estudios morfologicos en el Genero Narcissus L. Anatomia de hoja y escapa, III. Fontqueria 27:103–133
Ellul P, Boscaiu M, Vicente O, Moreno V, Rosello JA (2002) Intra- and interspecific variation in DNA content in Cistus (Cistaceae). Ann Bot 90:345–351
Engleheart GH (1894) Hybrid narcissi. J Royal Hort Soc 17:35–44
Fernandes A (1951) Sur la phylogenie des especes du genre Narcissus L. Bol Soc Brot II 25:113–190
Fernandes A (1959) On the origin of Narcissus cantabricus DC.. Bol Soc Brot 33:47–60
Fernandes A (1963) Sobre a evolucao do subgenero “Corbularia” do Genero ‘Narcissus’ L. Mem Acad Cienc Lisboa Cl Ciencia 8:363–381
Fernandes A (1966a) Nouvelles etudes caryologiques sur la section Jonquillae DC. du genre Narcissus. Bol Soc Brot II 40:207–261
Fernandes A (1966b) Le probleme de Narcissus tazetta L. Bol Soc Brot 40:277–319
Fernandes A (1968a) Contribution a la connaisance de la biosystematique de quelques especes du genre Narcissus L. Port Act Biol 9:1–44
Fernandes A (1968b) Keys to the identification of native and naturalized taxa of the genus Narcissus L. R.H.S Daffodil and Tulip Yearbook:37–66
Fernandes A (1975) L’evolution chez le genre Narcissus L. Anales Inst Bot Cavanilles 32:843–872
Fernandes A (1977) Sur la caryologie de Narcissus × hannibalis. Bol Soc Brot 51:201–206
Fernandes A (1991) Estudios cariologicos en Narcissus espanoles. Fontqueria 31:141–144
Fernandez Casas J (1977) Recuentos cromosomicos en plantas vasculares espanoles. Saussurea 8:33–55
Fernandez Casas J (1986a) Narcissus bujei. Lagascalia 14:176
Fernandez Casas J (1986b) Acerca de unos cuantos narcisos nortenos. Fontqueria 11:15–23
Fernandez Casas J (1991) Tres narcisos pirenaicos. Fontqueria 31:235–248
Fernandez Casas FJ (1996) Narcissorum notulae, II. Fontqueria 44:253–262
Fernandes A, Fernandes R (1946) Sur la caryo-systematique du sous-genre Ajax Spach du genre Narcissus L. Acta Univ Coimbrig 1–33
Fernandes A, Fernandes R (1951) On the origin of Tapeinanthus humilis. Herbertia 12:85–96
Fernandes A, Franca F (1974) Sur le comportement des heterochromatinosomes chez une population de Narcissus hispanicus Gouan. Bol Soc Brot 48:30–35
González-Aguilera JJ, Ludeña Reyes P, Fernández-Peralta AM (1990) Intra- and interspecific variations in nuclear parameters of two closely related species of Narcissus L.. Genetica 82:25–31
Graham SW, Barrett SCH (2004) Phylogenetic reconstruction of the evolution of stylar polymorphism in Narcissus (Amaryllidacae). Amer J Bot 91:1007–1021
Greilhuber J (1979) Evolutionary changes of DNA and heterochromatin amounts in the Scilla bifolia Group (Liliaceae). Pl Syst Evol Suppl 2:263–280
Greilhuber J (1995) Chromosomes of the monocotyledons. In: Rudall PJ, Cribb P, Cutler DF, Humphries CJ (eds) Monocotyledons: systematics and evolution. Kew, Royal Botanical Gardens, pp 379–414
Greilhuber J (1998) Intraspecific variation in genome size: a critical reassessment. Ann Bot 82:27–35
Greilhuber J (2005) Intraspecific variation in genome size in Angiosperms: identifying its existence. Ann Bot 95:91–98
Greilhuber J, Speta F (1978) Quantitative analysis of C-banded karyotypes and systematics in the cultivated species of the Scilla siberica group (Liliaceae). Pl Syst Evol 129:63–109
Haworth AH (1831) Narcissearum Monographia. London
Hirsch A, Testolin R, Brown S, Chat J, Fortune D, Bureau J, De Nay D (2004) Embryo rescue from interspecific crosses in the genus Actinidia. Pl Cell Rep 20:508–516
Hodgins KA, Barrett SCH (2007) Population structure and genetic diversity in tristylous Narcissus triandrus: insights from microsatellite and chloroplast DNA variation. Molec Ecol 16:2317–2322
Kalender R, Tanskanen J, Immonen S, Nevo E, Schulman AH (2000) Genome evolution of wild barley (Hordeum spontaneum) by Bare-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA 97:6603–6607
Leitch IJ, Chase MW, Bennett MD (1998) Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Ann Bot 82 (suppl A):85–94
Lifante ZD, Camacho CA (2007) Morphological variation of Narcissus serotinus s.l. (Amaryllidaceae) in the Iberian peninsula. Bot J Linn Soc 154:236–257
Maire R (1959) Narcissinae Link. Flore de l’Afrique du Nord 6:51–75
Mathew B (2002) Narcissus and daffodil classification of the genus Narcissus. In: Hanks R (ed) Narcissus and daffodils. Taylor & Francis, London, pp 31–52
Meadows GEM (1972) A host of daffodils. Q Bull Alp Gard Soc 40:98–106
Mol de WE (1923) The disappearance of diploid and triploid magnicoronate narcissi from the larger cultures and the appearance in their place of the tetraploid forms. Kon Acad Amst Proc Sect Sci 25:216–220
Montserrat Marti JM, Vives P (1991) Numeros cromosomaticos en Narcissi (Amaryllidaceae). Fontqueria 31:145–148
Nava Fernandez HS, Fernandez Casado A (1991) Narcisos hybridos del Noroeste Iberico. Fontqueria 31:249–254
Norman T, Norman M (2003) No title. Alp Gard 71:124–126
Nutt RD (1993) In: Beckett K (ed) Encyclopedia of Alpines 1. Pershore, Alpine Garden Society Publications, pp 510–520
Ohri D (1998) Genome size variation and plant systematics. Ann Bot 82 (suppl A):750–812
Perez-Barrales R, Vargas P, Arroyo J (2006) New evidence for the Darwinian hypothesis of heterostyly: breeding systems and pollinators in Narcissus section Apodanthi. New Phytol 171:553–567
Pugsley HW (1933) A monograph of Narcissus, subgenus Ajax. J Roy Hort Soc 58:17–93
RHS (2006) The international daffodil register and classified list. Royal Horticultural Society, London
Rios-Ruiz S, Rivera-Nunez D, Algaraz-Ariza F, Obon de Castro C (1998) Three new species of Narcissus L. subgenus Ajax Spach (Amaryllidaceae) restricted to the meadows and forests of south-eastern Spain. Bot J Linn Soc 131:153–165
Romero AT, Sachez Castillo PM, Ruiz Rejon M (1983) Sobre cariologia, morfologia et corologia de Narcissus tortifolius F. Casas. Fontqueria 4:7–10
Rouy G (1912) Flore de France 13:26–55
Sanudo A (1984) Estudios citogeneticos y evolutivos en poblaciones Espanoles del genero Narcissus L. sect. Pseudonarcissi DC. Nota previa: numeros de cromosomas. Anales Jard Bot Madrid 40:361–367
Smythies BE (1973) A note on Narcissus hispanicus Gouan (N. major Curtis) in the Sierra de las Nieves, Malaga Lagascalia 3:49–52
Susanna A, Garnatje T (2005) Some Narcissus from Northern Iberia. Curtis Bot Mag 22:124–138
Tiersch TR, Chandler RW, Wachtel SSM, Ellias S (1989) Reference standards for flow cytometry and application in comparative studies of nuclear DNA content. Cytometry 10:706–710
Thompson JD (2005) Plant evolution in the Mediterranean. Oxford University Press, Oxford
Uribe-Echebarria Diaz PM (1998) Sobre el gruppo Narcissus asturiensis–N. jacetanus en la peninsula Iberica. J Bot Soc France 5:147–154
Webb DA (1978) Taxonomic notes on Narcissus L. Bot J Linn Soc 76:297–307
Webb DA (1980) Narcissus. In: Tutin TG et al (eds) Flora Europaea 5:78–84 Cambridge
Wylie AP (1952) The history of the garden narcissi. Heredity 6:137–156
Yokoya K, Roberts AV, Mottley J, Lewis R, Brandham PE (2000) Nuclear DNA amounts in roses. Ann Bot 85:557–561
Zonneveld BJM (2001) Nuclear DNA contents of all species of Helleborus discriminate between species and sectional divisions. Pl Syst Evol 229:125–130
Zonneveld BJM, van Iren F (2001) Genome size and pollen viability as taxonomic criteria: application to the genus Hosta. Pl Biol 3:176–185
Zonneveld BJM, Grimshaw JM, Davis AP (2003a) The systematic value of nuclear DNA content in Galanthus. Pl Syst Evol 241:89–102
Zonneveld BJM, Duncan GD (2003b) Taxonomic implications of genome size and pollen colour and vitality for species of Agapanthus L’Heritier (Agapanthaceae). Pl Syst Evol 241:115–123
Zonneveld BJM, Duncan GD (2006) Genome size for species of Nerine Herb. (Amaryllidaceae) and its evident correlation with growth cycle, leaf width and other morphological characters. Pl Syst Evol 257:251–260
Acknowledgments
I like to thank for the contribution of plant materials by the donors listed. Special thanks go to John Blanchard and Derrick Donnison-Morgan who both provided most of the plant material and were always willing based on their extensive knowledge to discuss Narcissus matters with me. Dr. J.F. Veldkamp of the National Herbarium Leiden is thanked for going critically over the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Appendix: New combinations
Appendix: New combinations
N. jonquilla ssp. cordubensis (Fern. Casas) Zonn. comb. et stat. nov. Basionym: N. cordubensis Fern. Casas. (1982) Fontqueria 1: 10.
N. jonquilla ssp. fernandesii (Pedro) Zonn. comb. et stat. nov. Basionym: N. fernandesii Pedro (1947) Bol. Soc. Brot. II, 21: 60.
N. jonquilla ssp. willkommii (A. Fern.) Zonn. comb. et stat. nov. Basionym N. willkommii (Samp.) A. Fern. (1966) Bol. Soc. Brot. II, 40: 213.
N. longispathus ssp. alcaracensis (S. Rios, D. Rivera, Alcaraz & Obon) Zonn. comb. et stat. nov. Basionym: N. alcaracensis S. Rios, D. Rivera, Alcaraz & Obon. (1998) Bot. J. Linn. Soc 131: 153–165.
N. longispathus ssp. segurensis (S. Rios, D. Rivera, Alcaraz & Obon) Zonn. comb. et stat. nov. Basionym: N. segurensis S. Rios, D. Rivera, Alcaraz & Obon. (1998) Bot. J. Linn. Soc 131: 153–165.
N. longispathus ssp. yepesii (S. Rios, D. Rivera, Alcaraz & Obon) Zonn. comb. et stat. nov. Basionym: N. yepesii S. Rios, D. Rivera, Alcaraz & Obon. (1998) Bot. J. Linn. Soc 131: 153–165.
N. romieuxii ssp. jacquemoudii (Fern. Casas) Zonn. comb. et stat. nov. Basionym: N. jacquemoudii Fern. Casas (1986) Fontqueria 10: 11.
New taxa already mentioned in the text:
N. blanchardii Zonn. stat. nov.
Ancient allotetraploid form of N. jonquilla (ssp. fernandesii) × N. gaditanus
Based on N. fernandesii var. major A. Fern., Bol. Soc. Brot. II, 40: 239 (1966) Named after John Blanchard who wrote an eminent book on Narcissus.
Section Juncifolii (A. Fern.) Zonn. stat. nov. Typus N. assoanus Dufour ex Schult.& Schult.f in J. J. Roemer & J. A. Schultes, Syst. Veg. 7: 962 (1830). Based on subsection Juncifolii A. Fern. (1966). Bol. Soc. Brot. II, 40: 207–261. Species included: N. assoanus Dufour, N. gaditanus Boiss. & Reuter
Section Nevadensis Zonn. sect nov. Typus: N. nevadensis Pugsley, J. R. Hort. Soc. 58: 62 (1933). Scapus multiflores, corona and spatha longissima, DNA contentus nucleorum 30–39 pg a sectione Pseudonarcissus differt.
(Sub)species included: N. bujei Fern. Casas, N. longispathus ssp. alcaracensis (S. Rios, D. Rivera, Alcaraz & Obon) Zonn., N. longispathus ssp. longispathus Pugsley, N. longispathus ssp. segurensis (S. Rios, D. Rivera, Alcaraz & Obon) Zonn., N. longispathus ssp. yepesii (S. Rios, D. Rivera, Alcaraz & Obon) Zonn., N. nevadensis Pugsley.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Zonneveld, B.J.M. The systematic value of nuclear DNA content for all species of Narcissus L. (Amaryllidaceae). Plant Syst Evol 275, 109–132 (2008). https://doi.org/10.1007/s00606-008-0015-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-008-0015-1